Racial difference of mutational signature in hepatocellular carcinoma
Abstract
Aim: Previous studies have demonstrated the racial disparities of new incidence and mortality rate of hepatocellular carcinoma (HCC) patients, but the racial differences in the tumor characteristics causing these disparities remain unclear.
Methods: We collected genomic mutation profile of 589 HCC patients, including Asian-Korea (n = 231), Asian-TCGA (n = 156), White-TCGA (n = 176), and Black-TCGA (n = 16). We applied a non-negative factorized matrix algorithm to decipher the mutational signatures of HCC patients, compared racial differences of mutational signature, performed molecular subtyping analysis of HCC patients based on their composition of mutational signatures, and evaluated their influence on clinical outcome.
Results: Asian patients showed a significantly higher level of SBS96F-aristolochic acid exposure signature related to the widespread usage of Chinese herbs in East Asia, and they also showed higher SBS96B-MMR at T > C mutations but lower SBS96D-MMR at C > T mutations than White patients, suggesting the heterogeneous mechanisms related to defective DNA mismatch repair across races. Asian-Korea patients showed a significantly higher SBS96C-tobacco chewing and aflatoxin exposure than the other three populations, indicating the higher levels of aflatoxin contamination in food and environment in this area. The SBS96G-Unclear signature was also observed to be significantly higher in Asian-Korea patients, and their dominated subgroup patients showed better prognosis for both disease-free and overall survival probability.
Conclusion: Our study found racial differences of mutational signatures to be associated with differences in diverse genetic backgrounds and environmental factors, which might help guide the personalized treatment of HCC patients.
Keywords
INTRODUCTION
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is the fourth most common cause of cancer-related deaths worldwide[1]. Numerous epidemiologic studies demonstrated that the incidence rates and mortality of HCC patients varied by geographical region and socioeconomic status, such as high incidence in East Asia and Mongolia, the intermediate incidence rate in Europe and North America, and low incidence rate in South-Central Asia[2]. The incidence of HCC is closely associated to chronic viral hepatitis B or C infection and toxin exposure such as alcohol, aflatoxin, tobacco, or nonalcoholic steatohepatitis[3-5].
Over the past two decades, large-scale next-generation sequencing (whole genome and exome sequencing) has been performed in enormous studies to explore the genomic landscape of HCC, including recurrent driver mutations, copy number alterations, related dysfunctional biology pathways, and intratumor heterogeneity[4,6,7-12], which provided abundant resources to study the tumorigenesis of HCC. Somatic mutations in cancer genomes accumulated in mutational processes of both exogenous and endogenous factors occurring through the evolution history of cancer cell[13]. Each mutational process is reflective of the tumor intrinsic features including aging and mismatch despair deficiency and previous exposures to exogenous factors such as tobacco and drugs. In total, 94 single-base substitutions signatures with clear etiologies have been identified in previous pan-cancer study of over 30 cancer type[14]. The mutational signature analysis could provide evidence to build the association of exogenous and endogenous factor exposures with specific cancer risks or preferred treatments and further explain the epidemiology difference of HCC patients.
To study whether there is any racial difference of mutational signatures in HCC patients, we collected the genomic mutation profile of 589 HCC patients with different racial origins from previous studies[11,12], including 231 Asian-Korea HCC patients, 156 Asian-TCGA patients, 176 White-TCGA patients, and 16 Black-TCGA patients. Mutational signature extraction analysis was performed to compare their composition difference across races, and their influence on clinical outcomes was further evaluated.
METHODS
Data sources and clinical information
This study included 589 hepatocellular carcinoma patients in total [231 patients from Korea cohort[11] and 358 patients from The Cancer Genome Atlas (TCGA) cohort[12]]. Single nucleotide variants in mutation annotation format (MAF) from whole-exome sequencing and clinical information data, including survival time, race, and age, were downloaded from the cBioPortal website (https://www.cbioportal.org/datasets). Patients without clear race definition were removed, and the remaining patients were grouped into Asian-Korea, Asian-TCGA, White-TCGA, and Black-TCGA populations.
Mutation signature analysis
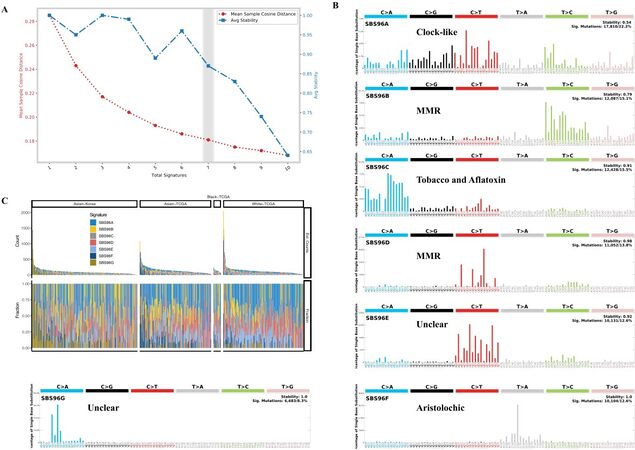
We applied the computational framework SigProfiler to decipher mutational signature profiles of 589 HCC patients and assign the contributions of each signature to each patient, based on the previously described methodology[14-17]. We fed the computational framework with the MAF profile of single nucleotide variants for all patients as input, and then ran the framework with two main steps: The first step, named SigProfilerExtraction, was based on somatic mutations in sequence context and their distributions in each patient, and it used multiple non-negative matrix factorization (NMF) iterations (10,000-1,000,000) to decipher a minimal set of mutational signatures that optimally explains the faction of each mutation context type in each mutational signature and estimate their activity for each sample. The second step, named SigProfilerAttribution, was responsible for accurately estimating the number of somatic mutations related to each extracted mutational signature in each patient. The optimal number of signatures as seven was determined by the trade-off of the mean sample cosine distance and average stability of solutions in the range from 1 to 10 [Figure 1A].
Figure 1. Deciphering mutational signatures of 589 hepatocellular carcinoma patients: (A) the distribution of error (mean sample cosine distance) and average stability of solutions for a range from 1 to 10; (B) mutational signatures identified by the SigProfiler; and (C) the estimated count and fractions of seven mutational signatures in HCC patients.
Proposed etiology annotation of mutational signatures
We used 94 known Catalogue of Somatic Mutations in Cancer (COSMIC) mutational signatures
De novo extraction of mutational signatures and their proposed etiology annotation
| De novo extracted | Global COSMIC signatures | Proposed etiology | Cosine similarity | Correlation |
| SBS96A | Signature SBS1 (5.26%, clock-like) & Signature SBS5 (34.14%, clock-like) & Signature SBS40 (60.60%, clock-like) | Clock-like | 0.85 | 0.65 |
| SBS96B | Signature SBS5 (48.80%, clock-like) & Signature SBS26 (51.20%, defective DNA mismatch repair) | MMR | 0.89 | 0.82 |
| SBS96C | Signature SBS5 (5.58%, clock-like) & Signature SBS24 (43.54%, aflatoxin exposure) & Signature SBS29 (50.88%, tobacco chewing) | Smoke and aflatoxin | 0.95 | 0.93 |
| SBS96D | Signature SBS1 (33.40%, clock-like) & Signature SBS5 (16.32%, clock-like) & Signature SBS15 (50.28%, defective DNA mismatch repair) | MMR | 0.97 | 0.96 |
| SBS96E | Signature SBS1 (1.06%, clock-like) & Signature SBS5 (30.76%, clock-like) & Signature SBS7a (7.02%, ultraviolet light exposure) & Signature SBS7b (11.12%, ultraviolet light exposure) & Signature SBS11 (27.66%, chemotherapy treatment) & Signature SBS23 (22.38%, unknown) | Unclear | 0.98 | 0.98 |
| SBS96F | Signature SBS1 (0.24%, clock-like) & Signature SBS5 (0.88%, clock-like) & Signature SBS22 (98.88%, aristolochic acid exposure) | Aristolochic | 0.88 | 0.87 |
| SBS96G | Signature SBS45 (37.68%, possible sequencing artefact) & Signature SBS49 (62.32%, possible sequencing artefact) | Unclear | 1 | 1 |
Unsupervised clustering of patients based on mutation signature components
Based on the contributions of signatures to each patient, we calculated their fraction among HCC patients to create a numerical matrix (patients as columns, signatures as rows) and then applied the NMF “lee” algorithm to perform molecular subtyping of HCC patients. After the manual inspection, we chose the K = 7 solution and reported seven subgroups.
Statistical analysis
Two-sided Mann-Whitney tests were performed using Wilcoxon signed-rank test to generate the empirical P values and plotted with the R package “ggpubr”.
Survival analysis
Chi-square test statistics were computed using log-rank tests, and Kaplan-Meier curves were plotted using the R package “survival” and “survminer”.
RESULTS
Identification of mutational signatures in HCC patients
Somatic mutations occur throughout life and display distinctive tumor cell-intrinsic patterns, including DNA replication machinery and defective DNA repair, as well as exogenous processes such as tobacco exposure and chemotherapy treatment. Different mutational processes generate distinct combinations of mutation types, which can be classified into 96 classes constituted by the six base substitutions C > A, C > G, C > T, T > A, T > C, and T > G. In total, 84,585 single nucleotide variants from 589 HCC patients were downloaded from a public database (cBioPortal) and categorized into a 96 mutation classes × 589 patient numeric matrix. We then applied SigProfiler software to extract optimal mutational signatures of HCC patients. Finally, the seven mutational signatures were identified in our study based on the trade-off curve of error and stability with the range from 1 to 10 [Figure 1A], which showed a distinct mutational pattern: SBS96A was characterized predominantly by C > T and C > G mutations; SBS96B was composed predominantly of T > C mutations; SBS96C was characterized primarily by C > A mutations at GCA and GCC trinucleotides (the mutated base is underlined); SBS96D was composed predominantly of C > T at ACG, CCG, and GCG trinucleotides; SBS96E was composed primarily of C > T at ACC, CCC, and TCC trinucleotides; SBS96F included enrichment of T > A mutations at CTG trinucleotides; and SBS96G was composed mainly of C > A at CCA and CCG trinucleotides [Figure 1B]. After dividing our study cohorts into four populations of Asian-Korea (n = 231), Asian-TCGA (n = 156), White-TCGA (n = 176), and Black-TCGA (n = 16), we observed that the seven mutational signatures showed diverse contributions to patients of the four different populations in the estimated count or composition fraction level [Figure 1C], indicating the mutational signature heterogeneity of HCC patients.
To investigate which etiology was proposed for the seven mutational signatures, we performed decomposition analysis with 94 known cosmic signatures [Table 1]. SBS96A was reconstructed with several different known clock-like COSMIC signatures (SBS1, 5.26%; SBS5, 34.14%; SBS40, 60.60%) and could be defined as a clock-like signature. SBS96B and SBS96D consisted mainly of cosmic signatures associated with defective DNA mismatch repair (MMR) and could be characterized as MMR-like signatures. SBS96C was also reconstructed by 43.54% aflatoxin exposure SBS24 and 50.88% tobacco chewing SBS29. SBS96F consisted of 98.88% SBS22, and could be defined as an aristolochic acid-exposure signature. SBS96E consisted of multiple COSMIC signatures related to clock-like, ultraviolet light exposure, and unknown factor. SBS96G consisted of signatures arising from possible sequencing artifacts [Table 1].
Racial difference of mutational signatures in HCC patients
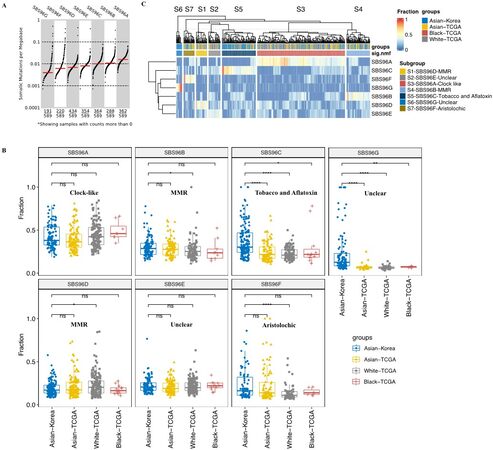
To explore whether there were any racial differences in mutational signatures, we compared the faction of seven mutation signatures among four populations, including Asian-Korea, Asian-TCGA, White-TCGA, and Black-TCGA. In general, the clock-like SBS96A signature was the major signature of HCC patients with the highest number of somatic mutations per megabase, while the SBS96G signatures as the minor signature showed the lowest activity but still existed in 32.4% of HCC patients (191/589) [Figure 2A]. SBS96A clock-like signature, related to the age of patients, and SBS96E signature, with unclear proposed etiology, showed no obvious difference among the four populations, suggesting that these two signatures were common in HCC patients [Figure 2B]. Compared with the Asian-Korea patients, the fractions of both SBS96B and SBS96D, as the two subsets of defective DNA MMR-related signatures, showed no significant changes in Black-TCGA and Asian-TCGA but showed adverse significant changes in White-TCGA (P < 0.05), indicating the heterogeneous mechanisms of defective DNA MMR in HCC patients [Figure 2B]. Interestingly, the fraction of SBS96F signature showed no obvious difference in the two Asian populations (Asian-Korea and Asian-TCGA) but was significantly lower in White-TCGA patients [Figure 2B]. The higher fraction of aristolochic acid-exposure signature in Asian patients might be caused by the usage of Chinese herbs that produce aristolochic acid[18-22]. Intriguingly, Asian-Korea populations showed a significantly higher fraction of SBS96C associated with tobacco chewing and aflatoxin exposure compared with the three populations from the TCGA cohort [Figure 2B], suggesting high levels of aflatoxin contamination in food and environment. It is also worth noting that the fraction of SBS96F signature arising from possible sequencing artifacts was observed to be significantly higher in the Asian-Korea cohort than in the other three populations from the TCGA cohort [Figure 2B]. These results demonstrate that the composition of mutational signatures was diverse across the HCC populations.
Figure 2. Racial differences in the mutational signatures of HCC. (A) The distribution of somatic mutations per megabase of signatures. (B) Comparison of prevalence of signatures among Asian-Korea, Asian-TCGA, White-TCGA, and Black-TCGA. Wilcoxon signed-rank test and Bonferroni corrected: P > 0.05, 0.01 < P ≤ 0.05, 0.001 < P ≤ 0.01, 0.0001 < P ≤ 0.001, and 0 < P ≤ 0.0001 are shown as ns, *, **, ***, and ****, respectively. (C) Unsupervised clustering of HCC patients based on the fractions of seven mutational signatures in each patient.
To investigate whether HCC patients could be characterized by the mutational signature components, we applied a non-negative factorized matrix algorithm on subtyping HCC patients based on the fraction change pattern of seven mutational signatures. We identified seven subgroups of HCC patients: S1-SBS96D-MMR dominated, S2-SBS96E-unclear dominated, S3-SBS96A-clock-like dominated, S4-SBS96B-MMR dominated, S5-SBS96C-tobacco and aflatoxin dominated, S6-SBS96G-unclear dominated, and S7-SBS96F-aristolochic dominated subgroup patients [Figure 2C]. Interestingly, S6-SBS96G-unclear and S5-SBS96C-tobacco, and aflatoxin exposure dominated patients were enriched in the Asian-Korea population [Figure 2C]. These results suggest that the dominant architectures of mutational signature across HCC patients are distinct and could be used to group HCC patients into heterogenous subtypes.
Prognostic analysis of distinct mutational signatures-dominant HCC subgroup patients
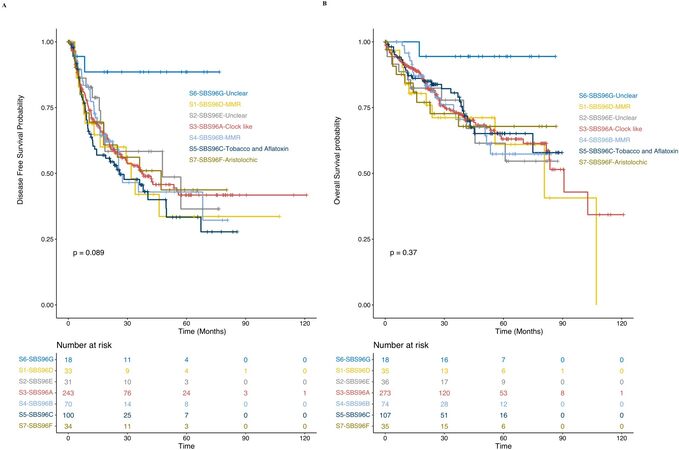
To explore whether there are different clinical outcomes of the seven subgroups of HCC patients characterized by specific dominant signature, we performed log-rank tests and Kaplan-Meier survival analyses of disease-free survival probability and overall survival probability. S5-SBS96C-tobacco and aflatoxin dominated subgroup showed worse prognosis in disease-free survival probability, while the S6-SBS96G-unclear dominated subgroup was observed to have favorable prognosis in disease-free and overall survival probability, despite the limited sample size [Figure 3]. More evidence from a larger cohort is needed to further exclude the potential of sample bias. These results demonstrate that the dominant architectures of a specific mutational signature caused by diverse genetic backgrounds or environmental factors has an important influence on HCC patients’ clinical outcomes, which could be used as biomarkers or predictors to guide personalized treatment of HCC patients.
DISCUSSION
Hepatocellular carcinoma is the primary type of liver cancer and is one of the most common malignant tumors in Asia. The genomic landscape of HCC was well-characterized by numerous previous studies, but their differences across races remain unclear. In this study, multiple significant racial differences of mutational signatures were demonstrated. We identified more prevalence of SBS96F-aristolochic acid exposure signature in HCC patients from Asia, which might be caused by the common usage of Chinese herbs in this area and is consistent with a previous study[18], suggesting that the detection and alternative usage of ingredients generating aristolochic acid will help reduce the incidence of HCC in Asia. We also showed a higher level of SBS96B-MMR at T > C mutations in Asian patients and a higher level of SBS96D-MMR at C > T mutations in White patients. MMR was defined as the predictor to sensitize cancer cells to immunotherapy response and showed high efficiency in the immune checkpoint blockade therapy of patients across many cancer types[23-25]. Differential enrichment of two signatures associated with defective DNA MMR mechanisms across races might explain why HCC patients showed a low response rate to immunotherapy and demonstrated the importance of classifying the MMR-like signature subtype before applying immunotherapy on patients with distinct racial origins.
Clustering of HCC patients based on their mutational signature exposures could stratify the patients into different prognostic subgroups. The HCC patients with the dominated prevalence of SBS96C-tobacco chewing and aflatoxin showed worse disease-free survival prognosis and were enriched in Asian-Korea patients, indicating that the level of aflatoxin contamination in food and environment was higher in Korea compared with North America, which might reflect the correlation of the mortality of HCC patients with the comprehensive socioeconomic status of a region. The SBS96G signature was also observed to be significantly higher in Asian-Korea patients, and their dominated subgroup patients showed better prognosis for both disease-free and overall survival probability, which might suggest the potential role of this signature to predict better prognosis of HCC patients. SBS96G signature was still observed in about 32.4% of HCC patients, and its etiology remained unclear. We also could not exclude the possibility of sequencing bias as the SBS96G signature could be reconstructed by two signatures generated by possible sequencing artifacts reported in a previous study[14].
Overall, our study identified multiple mutational signatures that varied substantially among different racial populations and evaluated their potential usage for predicting the clinical outcome of HCC patients. However, further validation in a larger cohort is needed before applying it to the personalized treatment of patients.
DECLARATIONS
Authors’ contributionsMade substantial contributions to conception and design of the study and performed data analysis and interpretation: Zhang BF, Guan XY
Wrote the draft and revised the manuscript: Zhang BF, Guan XY
Availability of data and materialsMutation and clinical information data could be obtained from the cBioPortal website
This work was supported by supported by grants from the Hong Kong Research Grant Council (RGC) including Collaborative Research Funds (C7065-18GF and C7026-18GF), Theme-based Research Scheme (T12-704/16-R), Shenzhen Science and Technology programs (KQTD20180411185028798), Shenzhen Fundamental Research Program (JCYJ20180508153249223). Professor Guan XY is Sophie YM Chan Professor in Cancer Research.
Conflicts of interestBoth authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
3. Akinyemiju T, Abera S, Ahmed M, et al. Global Burden of Disease Liver Cancer Collaboration. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol 2017;3:1683-91.
4. Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8.
5. Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018.
6. Marquardt JU, Seo D, Andersen JB, et al. Sequential transcriptome analysis of human liver cancer indicates late stage acquisition of malignant traits. J Hepatol 2014;60:346-53.
7. Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11.
8. Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226-39.e4.
9. Nault JC, Martin Y, Caruso S, et al. Clinical impact of genomic diversity from early to advanced hepatocellular carcinoma. Hepatology 2020;71:164-82.
10. Müller M, Bird TG, Nault JC. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol 2020;72:990-1002.
11. Ahn SM, Jang SJ, Shim JH, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60:1972-82.
12. Bailey MH, Tokheim C, Porta-Pardo E, et al. MC3 Working Group. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371-85.e18.
14. Alexandrov LB, Kim J, Haradhvala NJ, et al. PCAWG Mutational Signatures Working Group. The repertoire of mutational signatures in human cancer. Nature 2020;578:94-101.
15. Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246-59.
16. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Australian Pancreatic Cancer Genome Initiative. Signatures of mutational processes in human cancer. Nature 2013;500:415-21.
17. Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52-60.
18. Ng AWT, Poon SL, Huang MN, et al. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med 2017;9:eaan6446.
19. Lu ZN, Luo Q, Zhao LN, et al. The mutational features of aristolochic acid-induced mouse and human liver cancers. Hepatology 2020;71:929-42.
20. Mei N, Arlt VM, Phillips DH, Heflich RH, Chen T. DNA adduct formation and mutation induction by aristolochic acid in rat kidney and liver. Mutat Res 2006;602:83-91.
21. Hu SL, Zhang HQ, Chan K, Mei QX. Studies on the toxicity of Aristolochia manshuriensis (Guanmuton). Toxicology 2004;198:195-201.
22. Chang SY, Weber EJ, Sidorenko VS, et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2017;2:95978.
23. Chalabi M, Fanchi LF, Dijkstra KK, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med 2020;26:566-76.
24. Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol 2018;36:773-9.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Zhang BF, Guan XY. Racial difference of mutational signature in hepatocellular carcinoma. Hepatoma Res 2021;7:62. http://dx.doi.org/10.20517/2394-5079.2021.81
AMA Style
Zhang BF, Guan XY. Racial difference of mutational signature in hepatocellular carcinoma. Hepatoma Research. 2021; 7: 62. http://dx.doi.org/10.20517/2394-5079.2021.81
Chicago/Turabian Style
Zhang, Bai-Feng, Xin-Yuan Guan. 2021. "Racial difference of mutational signature in hepatocellular carcinoma" Hepatoma Research. 7: 62. http://dx.doi.org/10.20517/2394-5079.2021.81
ACS Style
Zhang, B.F.; Guan X.Y. Racial difference of mutational signature in hepatocellular carcinoma. Hepatoma. Res. 2021, 7, 62. http://dx.doi.org/10.20517/2394-5079.2021.81
About This Article
Copyright
Data & Comments
Data
 Cite This Article 9 clicks
Cite This Article 9 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.