Old-fashioned and newly discovered biomarkers: the future of NAFLD-related HCC screening and monitoring
Abstract
Nonalcoholic fatty liver disease (NAFLD) is the major contributor to the global burden of chronic liver diseases and ranges from simple and reversible steatosis to nonalcoholic steatohepatitis (NASH), which may progress into cirrhosis and hepatocellular carcinoma (HCC). HCC represents the most common liver cancer, and it is a leading cause of death worldwide with an increasing trend for the future. Due to late diagnosis, non-responsiveness to systemic therapy, and high cancer heterogeneity, the treatment of this malignancy is challenging. To date, liver biopsy and ultrasound (US) are the gold standard procedures for HCC diagnosis and surveillance, although they are not suitable for mass screening. Therefore, it is impelling to find new, less invasive diagnostic strategies able to detect HCC at an early stage as well as monitor tumor progression and recurrence. Common and rare inherited variations that boost the switching from NASH to liver cancer may help to predict tumor onset. Furthermore, epigenetic changes which reflect intertumoral heterogeneity occur early in tumorigenesis and are highly stable under pathologic conditions. The severity of hepatic injuries can be detected through the analysis of cell circulating tumor DNAs (ctDNAs), microRNAs (miRNAs), and noncoding RNAs (ncRNAs), which are involved in several pathological processes that feature cancer, including cell growth, survival, and differentiation, thus representing appealing biomarkers for HCC. Therefore, this review discusses the current options for HCC surveillance, focusing on the role of genetic and epigenetic biomarkers as new strategies to refine HCC management.
Keywords
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) constitutes the major contributor to the burden of chronic liver diseases, with a prevalence of approximately 25% in the general population. NAFLD encompasses a spectrum of histological conditions, ranging from simple steatosis to nonalcoholic steatohepatitis (NASH)[1,2]. The latter is characterized not only by triglyceride (TG) accumulation into the hepatic parenchyma but also by lobular inflammation and ballooning, and it may progress to fibrosis, cirrhosis, and hepatocellular carcinoma (HCC)[3], boosted by metabolic (obesity, type 2 diabetes (T2D), and insulin resistance (IR)), genetic, and epigenetic risk factors[4].
HCC may also develop in the absence of cirrhosis, although it is most common in patients with advanced fibrosis[5]. Indeed, NAFLD-related HCC reports an annual incidence in the USA and Europe that ranges from 0.7%-2.6% in patients with NASH-related cirrhosis and 0.1%-1.3% in non-cirrhotic ones[6].
Currently, the most effective therapy systemically administered to HCC patients is the multiple kinase inhibitor sorafenib[8]. Other therapeutic options involve invasive surgical approaches, such as partial resection of the liver, image-guided tumor ablation, and liver transplantation[9]. Nevertheless, the mechanisms underlying the development of HCC in the context of NAFLD, especially in the absence of cirrhosis, are not completely clarified. Thus, the identification of non-invasive biomarkers and possible therapeutic targets is crucial to improve HCC surveillance, diagnosis, and treatment, as well as avoid surgical interventions whenever possible. The present review briefly discusses the potential clinical utility of currently exploited non-invasive biomarkers and that of the newly discovered inherited or acquired ones in the screening and monitoring of NAFLD-driven HCC.
COMMON AND RARE GENETIC BIOMARKERS OF NAFLD-RELATED HCC
Genetics of HCC: how common variants may influence unfavorable outcomes
Increasing evidence and epidemiological studies indicate that NAFLD pathogenesis and transition from NASH to HCC have a strong heritable component, involving both common and rare mutations[10]. The rs738409 C > G single nucleotide polymorphism (SNP) in the patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene, namely PNPLA3 I148M, is strongly associated with the entire spectrum of NAFLD until HCC[11]. Patients carrying the at-risk G allele lose the PNPLA3 enzymatic activity, which drives TG accumulation in hepatocytes and predisposes to macro- and micro-vesicular steatosis, portal inflammation, and high proliferation of hepatic progenitor cells (HPCs)[12,13]. Moreover, PNPLA3 loss-of-function may trigger fibrogenesis and carcinogenesis, irrespective of steatosis[6]. Another important genetic contributor to NAFLD is the rs58542926 C > T (E167K) missense variant in the transmembrane 6 superfamily member 2 (TM6SF2) gene. The TM6SF2-encoded protein participates in hepatic very low-density lipoprotein (VLDL) lipidation and assembly in the endoplasmic reticulum (ER) cisternae. Notably, the rs58542926 variation leads to a misfolded protein, which is rapidly degraded in hepatocytes and leads to impaired VLDL secretion and fat accumulation[14]. Multicenter studies have confirmed that the TM6SF2 E167K variation in NAFLD patients is associated with lower serum cholesterol and TG levels, conferring protection against cardiovascular events[15]. Nevertheless, this variant was also associated with liver damage and advanced hepatic fibrosis/cirrhosis in several cohorts of patients[16-19]. However, its impact on HCC onset is still debated. Finally, the rs641738 C > T variant in the TMC4/MBOAT7 locus, also known as
Rare variants participating in NAFLD-HCC onset
Loss-of-function mutations in telomerase reverse transcriptase (TERT) were associated with familial cirrhosis and the predisposition to HCC onset[26,27]. Indeed, an exome sequencing analysis performed on NAFLD-HCC tumor samples revealed that mutations in TERT promoter contribute to NAFLD-related liver carcinogenesis. In detail, in the ten patients, TERT promoter mutations occurred along with catenin beta 1
Likewise, mutations in apolipoprotein B (APOB) gene, which regulates VLDL assembly and secretion, were correlated with severe hepatic fat deposition and liver injuries up to HCC[29]. Decreased lipoprotein levels have also been associated with a reduced risk of cardiovascular complications in HCC patients[30]. Similarly, the new rs599839 A > G variant in PSRC1, which belong to the CELSR2-PSRC1-SORT1 gene cluster that was demonstrated to be protective against dyslipidemia, was positively associated with cell proliferation in NAFLD patients. In a cohort of NAFLD-HCC (n = 131), the rs599839 variant was reported with higher frequency compared to NAFLD patients without cancer (n = 1295). In addition, the presence of this variant correlates with an increased HCC risk, independently of fibrosis severity, poor prognosis, and advanced tumor stage[31]. Finally, the rs1800832 A > G variant in the 5′ UTR of the neurotensin (NTS) gene has been shown to be associated with hepatic fibrosis, cirrhosis, and HCC in a large cohort of NAFLD patients
Polygenic risk scores, a strategy to predict disease prognosis
In the past years, candidate genes and genome-wide association studies (GWAS) have been exploited to evaluate the impact of every single genetic variant on the risk of developing severe NAFLD. However, it is worthwhile to aggregate individual genetic predictors into polygenic risk scores (PRSs) to foresee the diagnosis and prognosis of advanced NAFLD and to stratify patients according to their risk of progression towards HCC[33]. To date, several studies have tried to build reliable PRSs in NAFLD setting by using regression models or more complex statistical tools, correlating clinically relevant genetic variants with environmental and dynamic risk factors[10]. With this purpose, Dongiovanni et al. exploited in an elegant work a mendelian randomization analysis and PRS to evaluate the impact of the co-presence of PNPLA3, TM6SF2, MBOAT7, and GCKR at-risk alleles on progressive liver damage[34]. They demonstrated that the presence of these four at-risk alleles was directly associated with hepatic fat deposition in NAFLD patients, and the latter is causally implicated in the transition to severe chronic liver disease[34]. Thus, the impact of each genetic variant on HCC was directly proportional to the predisposition to fatty liver[35]. Similarly, Di Costanzo and colleagues highlighted the predominant role of the genetic factors PNPLA3, TM6SF2, and GCKR in determining the amount of liver fat content in children with obesity. They generated a weighted genetic risk score demonstrating that the three risk alleles more tightly participate in fat deposition. Indeed, PNPLA3, TM6SF2, and GCKR variants significantly increase the risk of NAFLD compared to IR, the predominant trigger factor of steatosis and fibrosis[36]. The cumulative effect of multiple genetic variants on NAFLD and HCC pathogenesis has been largely investigated. In particular, the combined effect of PNPLA3 and TM6SF2 mutations seems to have a predominant effect on lipid metabolism dysregulation and NAFLD progression[37-39]. The increased number of at-risk alleles in NAFLD and HCC patients was further associated with increased alanine aminotransferase (ALT), aspartate aminotransferase (AST), and
Finally, in a cohort of 1380 patients with NAFLD, among whom 121 had HCC, the impact of the three variants I148M PNPLA3, rs641738 MBOAT7, and E167K TM6SF2 was evaluated[41]. The co-presence of the three risk variants was correlated with increased levels of liver damage markers, higher grade of steatosis, lobular inflammation, ballooning, fibrosis, and approximately two-fold higher risk of developing HCC. Indeed, the prevalence of the three risk variants was 2.5-fold higher in patients with HCC compared to NAFLD, confirming that these variants predispose to NAFLD progression toward cancer[41].
In summary, PRSs combine the impact of different genetic variants in a prediction model which estimates the risk of developing liver disease. A “good” PRS combines the effect of different known risk alleles with clinical pathological traits, acquiring a more pronounced accuracy for the early detection of HCC[35]. Therefore, PRSs integrating more risk factors may be a useful strategy to predict disease prognosis.
WELL-ESTABLISHED AND NEWLY DISCOVERED CIRCULATING/TISSUE BIOMARKERS FOR HCC SURVEILLANCE
During tumor progression, liver cells are likely to present different molecular signatures. The latter, if properly assessed, could be potential biomarkers for hepatocarcinoma diagnosis and follow-up. Several different biomarkers are currently used in clinical practice for tumor staging, grading, and management, but no trustworthy biochemical dynamic parameter can precisely predict the progression of NASH towards HCC[42]. In this context, studies focused on the identification of novel non-invasive biomarkers with a reliable diagnostic power may be attractive, aiming to improve the early diagnosis and prognosis of HCC. To date, proteins that are highly expressed in HCC compared to adjacent non-tumoral tissues are the most appealing parameters for HCC surveillance, and, despite several limitations, they still have a high consensus among clinicians. Among them, alpha-fetoprotein (AFP), des-gamma-carboxy prothrombin (DCP), and glypican 3 (GPC-3) have the highest diagnostic performance for the detection of HCC[43-45].
Alpha-fetoprotein
AFP is the most widely used non-invasive biomarker for the diagnosis and prognosis of HCC. Blood levels of AFP were found to increase during chronic liver disease, in cirrhotic patients, and in HCC settings[46]. In a nested case-control study, increased AFP levels were observed six months before the diagnosis of HCC, suggesting a possible predictive role of this molecule. However, enhanced AFP concentrations are not exclusive to HCC, and they can also be observed in patients without tumors but with liver cirrhosis or chronic liver disease[47,48]. Moreover, the ability of AFP to detect HCC showed relatively good sensitivity but poor specificity, limiting its use in clinical practice[49]. This led the American Association for the Study of Liver Diseases (AASLD) Practice Guidelines Committee to recommend the use of ultrasound (US) alone for HCC surveillance[50]. For these reasons, AFP cannot be used as a single biomarker for HCC screening but, when combined with other strategies, can gain sensitivity[51]. For this purpose, a meta-analysis compared the efficacy of US with or without AFP for early HCC detection (n = 2770). The assessment of AFP in combination with abdominal US better detected early HCC development with a significant improvement in sensitivity (63% AFP combined with abdominal US vs. 45% US alone)[52].
AFP-L3, an AFP isoform more specific for malignant tumors, has emerged as a possible alternative strategy to monitor HCC progression and metastasis. Accordingly, the US Food and Drug Administration (FDA) recently approved the use of AFP-L3 to identify patients with a high HCC risk, considering the better specificity compared to AFP[48,53].
Des-gamma-carboxy prothrombin
Among the principal serological biomarkers approved by the FDA for HCC screening, DCP plays an important role in predicting HCC and, when combined with AFP, may complement US in the detection of early HCC[45]. DCP is an abnormal prothrombin molecule induced by vitamin K and produced during the malignant transformation of hepatocytes, and its overproduction during HCC suggests DCP suitability as a tumor prediction factor[43]. The association between DCP and liver cancer was firstly observed in a cohort of HCC (n = 1377) and chronic hepatitis or cirrhosis (n = 355) patients. Here, the ability of DCP and AFP to discriminate HCC from chronic liver diseases was tested, and DCP was demonstrated to better detect large tumors compared to the latter[54]. New evidence indicates that DCP could also be an appealing marker to discriminate advanced tumor stage, and it is currently used in clinics as a prognostic factor after therapy due to its involvement in the crosstalk between tumor and vascular endothelial cells[55]. Furthermore, DCP-positive and AFP-negative tumors show greater aggressiveness, larger size, less differentiation and vascular invasion, and recurrence after treatments. Thus, DCP may help to bridge gaps in the screening of more aggressive tumorigenic phenotypes[44]. However, even if DCP shows high diagnostic efficacy in certain conditions, it is not suitable for early HCC detection, and as a single biomarker, it does not offer substantial advantages compared to AFP[43,45].
The combined use of DCP, AFP, and AFP-L3 may refine HCC detection and has been recommended for clinical practice[49-51]. Indeed, AFP, DCP, and AFP-L3 have been merged in a serum biomarker-based model, “GALAD”, which also includes gender and age. GALAD in NAFLD patients was found to predict the probability of developing NASH and HCC with more sensitivity compared to both US scanning and all serum biomarkers alone. Hence, the GALAD score may be a reliable tool for HCC surveillance and may be exploited to identify patients at higher risk of NASH and HCC[56].
Glypican 3
A novel biomarker, GPC3, was strongly associated with tumor growth, and it also was recently proposed as a potential target for the diagnosis and treatment of HCC[57]. GPC3 is a heparin sulfate proteoglycan that interacts with several growth factors by binding to cell membrane via glycosylphosphatidylinositol anchors[58]. Increased levels of GPC3 were found in sera of HCC patients but not in healthy individuals or patients with hepatitis. Notably, Liu et al. demonstrated that the GPC3-positive rate was different in cirrhotic livers with or without HCC. In particular, cirrhotic livers with HCC were characterized by a higher GPC3-positive rate compared to those without tumors. In addition, GPC3 positivity increases HCC recurrence in cirrhotic HCC livers, suggesting the use of GPC3 as a precancerous biomarker[59]. In another study, elevated levels of GPC3 in tumor cells were related to poor prognosis with reduced five-year survival (54.5% vs. 87.7%; P = 0.031) compared to GPC3-negative ones. The soluble NH2-terminal fragment of GPC3 was used as a serological biomarker independently related to both overall survival (OS) (P < 0.05) and disease-free survival (DFS) (P < 0.01) and discriminates patients with small, well-differentiated HCC tumors and those with cirrhosis. In addition, GPC3 was recently recognized as an independent prognostic factor for DFS, indicating that GPC3 positivity may contribute to tumor recurrence after resection[60]. Although GPC3 concentrations might be indicative of the presence of HCC, similar to AFP, it suffers from low sensitivity and poor detection rate in blood samples compared to tissue biopsies. For these reasons, GPC3 does not meet the requirements of clinical practice as a single marker, but when combined with AFP gains in sensitivity and specificity[60].
Considering its potential, innovative GPC3-based therapeutic strategies have been proposed in clinical trials. Recently, a double-blind, phase II trial tested codrituzumab, a newly developed humanized monoclonal antibody against GPC3, in a cohort of 185 patients with HCC stratified according to GPC3 immunohistochemical expression. The drug, which is supposed to interact with CD16/FcγRIIIa and trigger antibody-dependent cytotoxicity, was administered to patients with advanced HCC who had failed prior systemic therapies. Unfortunately, codrituzumab did not show clinical benefit in treated patients. Nevertheless, the authors indicated that a higher dose of codrituzumab or the selection of patients with increased levels of GPC3 and CD16 in circulating immune cells may improve tumor outcomes[61].
Furthermore, 32A9a, a novel monoclonal antibody targeting the middle region of GPC3, was tested in mice with encouraging results. The antibody was administered in combined therapy with immunotoxin or chimeric antigen receptor T (CAR-T) designed against GPC3. The treatment promoted the regression of HCC in the tested models[62]. Other studies reported that GPC3-targeted CAR-T cells effectively destroyed GPC3+ HCC cells in vitro and tumor xenografts in mice. In addition, GPC3 CAR-T combined with sorafenib proved to be more effective against tumor growth[63]. Therefore, GPC3 can discriminate precancerous lesions in cirrhotic livers, and given its exceptional specificity, it may be used both as a novel therapeutic target and as a prognostic biomarker in HCC.
G protein-coupled receptors
Many G protein-coupled receptors (GPCRs), an important class of cell surface receptors involved in oncogenic transformation and tumor cell growth, have been reported to be mutated and overexpressed in tumor microenvironments, including HCC; thus, they are currently under evaluation as possible promising tumor biomarkers. In one study, GPCRs such as the beta2-adrenergic receptor (Beta2-AR), which initiates multiple signaling cascades and regulates cell proliferation through a classical cyclic-adenosine-monophosphate (cAMP)/protein kinase A (PKA) pathway, was found upregulated in HCC tumor tissues and in five HCC cell lines, exerting a role on tumor outcome[64]. The authors proposed Beta2-AR as a novel predictive factor for both recurrence-free survival and OS considering the significant association with poor prognosis independent of AFP, tumor-node-metastasis stage, and Edmondson stage[64]. Another study investigated the role of G-protein signaling modulator 2 during HCC carcinogenesis. This G protein receptor was enriched in HCC and hepatoma cell lines compared to healthy controls and was directly correlated with tumor size. In vitro experiments further corroborated that G-protein signaling modulator 2 accelerates cell growth, cell cycle, migration, and invasion and inhibits apoptosis. Indeed, it has the role of an oncogene in HCC through the activation of the phosphatidylinositol 3-kinase/protein kinase B signaling pathway[65]. Overall, these findings point out G-protein signaling modulator 2 as another attractive potential diagnostic biomarker to detect HCC. Accordingly, Saha et al. established GPR50 as a new appealing molecule to foresee HCC prognosis[66]. GPR50 is enhanced in HCC and forms a molecular complex with a disintegrin and metalloproteinase metallopeptidase domain 17 (ADAM17), regulating its activity and consequently Notch signaling pathway activation. As proof, GPR50 knockdown led to the suppression of Notch signaling and lowered tumor progression[66]. Interestingly, Gα12, a G protein that facilitates potent neoplastic transformation and change of cancer cells to a more aggressive phenotype, was found overexpressed in patients with HCC and correlated with the repression of liver-specific microRNAs, including miR-122, miR-148, miR-192, and miR-194[67].
Promising biomarkers in the era of immunotherapy for HCC
Dysregulation of immune hepatic milieu promoted by pathological metabolic changes is directly involved in tissue-damage processes and the evolution from NAFLD towards HCC[68]. As a consequence, several immunotherapeutic approaches have become attractive, and many efforts have been addressed to investigate the possibility of exploiting immunotherapy as the new standard of care in the management of HCC[69]. In particular, therapeutic strategies focused on immune checkpoint blockade (ICB) have been introduced, improving HCC prognosis[70]. Accordingly, ICB has been proposed as a useful tool to restore immune control of tumors by disrupting co-inhibitory receptors and enhancing anti-tumor T-cell responses. In particular, several regulatory pathways able to counteract immune system activation have been pinpointed as possible targets for immunotherapy, such as programmed cell death protein 1 (PD-1), a membrane protein expressed on all T cells[68], and cytotoxic T lymphocyte-associated protein 4 (CTLA-4), which competes with CD28 for binding of B7 molecules[71], both exerting a negative effect on T-cell proliferation and survival. Although immunotherapy is emerging as a promising strategy, reliable systems to detect its efficacy are required, and novel biomarkers have been proposed for this purpose. Among them, the increase in tumor-infiltrating lymphocytes (TILs), T cells infiltrating HCC, has been indicated as a marker of better response to therapy. In particular, CD3+ infiltrating T cells have been correlated with improved OS and response to nivolumab[72], whereas CD8+ enhancement showed clinical benefits for HCC in therapy with tremelimumab post ablation[73]. Similar results have been obtained considering TGF-β signaling activation as an indicator of exhausted immune signature in HCC. In this regard, HCC patients treated with pembrolizumab and with TGF-β levels < 200 pg/mL reported better OS and PFS, indicating TGF-β as a negative predictive biomarker for immune checkpoint inhibitor therapy[74]. Rapid real-time information regarding the prediction of treatment response and disease monitoring is also provided by the investigation of circulating tumor cells (CTCs), extracellular vesicles, and nucleic acids released[75]. These aspects are more extensively discussed in the following paragraphs.
GENE–ENVIRONMENT INTERACTIONS: NOVEL INSIGHTS INTO EPIGENETIC BIOMARKERS TO DETECT HCC ONSET
The evolution of NASH to HCC is characterized by multiple genetic and epigenetic alterations that run through cancer initiation, promotion, and progression[76]. Epigenetic modifications are hereditable but reversible phenomena that may dysregulate gene expression without modifying DNA sequence. These events may target DNA at different stages and include the substitution of DNA nucleotides (e.g., methylation), acetylation/deacetylation of histones that alters DNA packing and accessibility, or regulation of transcription that affects mRNA stability[76]. Recent studies have highlighted the potential of these novel biomarkers in NAFLD-HCC setting, building steps toward their clinical application. In this regard, activation of DNA methyltransferases (DNMTs) was significantly enhanced in NASH patients compared to those with simple steatosis, resulting in a higher methylation pattern of specific genes, including the mitochondrially encoded NADH dehydrogenase 6 (MT-ND6)[77]. This suggests that epigenetic changes in mtDNA may participate in the switching from simple steatosis to NASH. Afterwards, Kuramoto et al. determined the specific DNA methylation motif that in NASH-related tissues favors the silencing of genes implicated in the repair of damaged DNA and apoptosis, exacerbating the process of hepatocarcinogenesis. Notably, variations in DNA packaging due to post-translational epigenetic histone modifications were found to dysregulate the chromatin organization in NASH-related HCC[78]. The activation of histone deacetylase 8 (HDAC8) both in NAFLD-HCC patients and in rodents was found to be regulated by sterol regulatory element binding transcription factor 1 (SREBP1) and favors HCC development by inhibiting p53/p21-mediated apoptosis and cell-cycle arrest and stimulating Wnt/β-catenin signaling[79].
PROMISING TARGETS TO DIAGNOSE HCC: CELL-CIRCULATING TUMOR DNA AND NONCODING RNAS
To date, the characterization of the HCC molecular signature has significantly increased our ability to look into the tumor molecular pathogenesis and heterogeneity, unraveling new potential biomarkers. Liquid biopsy, intended as body fluids sampling, has recently emerged as a promising and innovative strategy aiming to assess the genetic profile of all cancerous lesions (primary and metastatic tumors) and dynamically track genomic tumor evolution[80]. The possibility of analyzing molecules released into the circulation is more feasible compared to invasive screening methods and may significantly improve tumor surveillance, detection, and outcome. A novel possible biomarker that is deemed to have great potential in HCC management is cell circulating tumor DNAs (ctDNAs). They are double-stranded DNA, found in plasma or serum, which contain genetic defects identical to the tumor cells from which they originated[81]. These DNA fragments showed hotspot genetic and epigenetic signatures that were correlated in many cancer types, including HCC, with tumor progression[82-84]. Likewise, growing evidence demonstrates that the dysregulation of noncoding RNAs (ncRNAs) expression is also associated with disease progression in cancers[85]. This class of molecules, including microRNA (miRNA), long noncoding RNA (lncRNA), and circular RNA (circRNA), does not encode proteins but still influences gene expression. In the HCC setting, various ncRNAs were found altered during tumor initiation, progression, and metastasis, exerting a possible regulatory role on NAFLD progression to HCC[86]. As biomarkers for cancer diagnosis and prognosis, HCC-related ncRNAs can be extracted from tumor tissues, peripheral blood, and urine samples of patients[87], highlighting their potential use as new non-invasive factors for HCC surveillance.
Cell circulating tumor DNA (ctDNA)
ctDNAs are derived from apoptotic and necrotic tumor cells that release their fragmented DNA into the circulation. ctDNAs possess specific cancer-associated molecular characteristics, such as single-nucleotide polymorphisms and methylation changes, that were found to have a significant role in determining cancer metastatic potential and recurrence[88]. Indeed, the “methylation pattern” of ctDNAs has been widely investigated in HCC and correlated with early tumorigenesis[89]. However, ctDNAs also play an important role in HCC prognosis, being an independent risk factor that is negatively associated with DFS, OS, recurrence, and extrahepatic metastasis[90]. Several methylation patterns have been associated with HCC, recognizing hypermethylation in promoter regions as an important early event in carcinogenesis. In addition, when combined with AFP, ctDNA detection can improve HCC diagnosis. As proof of this concept, a study demonstrated the elevated diagnostic value of ctDNA and AFP combination with 95.1% sensitivity and 94.4% specificity in discriminating HCC patients from healthy controls[91,92].
In solid tumors, ctDNA concentrations were still correlated with DFS, OS, and clinical response to ICB therapy with pembrolizumab, a monoclonal anti-PD-1 antibody, suggesting a potential clinical role as a predictive factor for ICB[93]. In the context of HCC, immunotherapy with PD-1 inhibitors has been tested in clinical trials against sorafenib-resistant tumors, and it is currently emerging as a new clinical option[68]. Indeed, a recent prospective phase II clinical trial assessed ctDNA levels in patients with advanced solid tumors treated with pembrolizumab, including HCC. Interestingly, changes in ctDNA levels from baseline were predictive of response to therapy, and they were also correlated with improved OS and PFS[93].
In keeping with these findings, several studies demonstrated that elevated levels of the PD-1 ligand (PD-L1) also correlate with poorer OS and worse clinical outcomes in patients with severe HCC, but it would not affect treatment response to immune checkpoint inhibitors, as monotherapy or in combination with other agents[94]. In a cohort of 48 unresectable HCC patients treated with atezolizumab, an anti-PD-L1 agent, plus bevacizumab, higher baseline concentrations of ctDNAs were associated with greater tumor burden, whereas dynamic changes in ctDNA levels post-treatment were associated with response to therapy[95]. Therefore, ctDNA evaluation has gained great diagnostic value in HCC, exceeding the previously described plasma biomarkers in terms of higher sensitivity and better clinical correlation. Similar to ctDNAs, the tumor mutational burden (TMB), defined as the overall number of somatic non-synonymous mutations per megabase in cancer cells, has also been proposed to identify HCC patients eligible for immunotherapy with PD-1 inhibitors[69] Even though TMB is generally low in HCC, its rise correlates with higher response rates to PD-1 inhibitors, and, among driver genetic alterations identified in HCC, the presence of activating WNT/β-actin mutation was associated with innate resistance to immune checkpoint inhibitors[96,97].
Short noncoding RNAs: microRNAs
miRNAs are endogenous, small noncoding RNAs that function in the regulation of gene expression[98]. miRNAs may work as either oncogenes or tumor suppressors under certain conditions; thus, they have been identified as potential biomarkers for cancer diagnosis and prognosis as well as therapeutic targets. Their dysregulation significantly contributes to liver disease progression to HCC[99]. For instance, a reduction in miR-122 was indicated to be a direct inducer of NASH-associated HCC[100]. Again, miR-15 and miR-16, which exert a tumor suppressor role in physiological conditions, were found downregulated in invasive HCC cell lines and aggressive HCC with lymph nodes metastasis and elevated TNM classification. Furthermore, hepatoma cells and tumor samples showed shortened expression of miR-34a, able to exert an anti-tumor activity via the p53/miR-34a/SIRT1 positive feedback loop[101]. On the contrary, the upregulation of other miRNAs, such as miR-221 and miR-101-3p, turned out to stimulate cell growth and invasion in cultured cells and in vivo models. Overall, miRNAs correlate with tumor prognosis and resistance to therapy in HCC patients and can be targeted by specific therapeutic agents[102].
Long noncoding RNAs
lncRNAs are molecules with a length of more than 200 nucleotides, and they work as transcriptional regulators both in the nucleus and the cytoplasm. Global understanding of lncRNA function is still pending, but recent studies reported their involvement in tumorigenicity and cancer progression through competitive endogenous RNA (ceRNA)-mediated regulatory mechanisms[85]. Several lncRNAs are involved in gene activation or silencing and are associated with chromatin modification enzymes. A small group of lncRNAs has recently been described as post-transcriptional regulators and could mediate mRNA processing (splicing), taking part in sequestering miRNAs away from their targets[103]. In the NAFLD-HCC setting, lncRNAs are likely to influence the susceptibility to liver disease with a potential role in the pathogenesis of HCC[104].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)
lncRNA MALAT1 plays a key role in tumor cell proliferation, migration, and invasion, and it seems to be involved in hepatic steatosis onset[104]. A recent study demonstrated that MALAT1 and lncRNA highly upregulated in liver cancer (HULC) were increased in fibrotic liver biopsies of patients with NAFLD, identifying C-X-C motif chemokine ligand 5 (CXCL5) as the possible target protein of MALAT1[105]. Fibrotic mice treated with CCl4 exhibited MALAT1 overexpression in fibrotic hepatic tissues with activation of hepatic stellate cells (HSCs)[106]. As a consequence, the knockdown of MALAT1 alleviated liver fibrosis in mice through the reduction of actin alpha 2 (Acta2) and collagen type I alpha 1 chain (Col1a1) levels. In HepG2 cells and T2D animal models, MALAT1 expression promoted hepatic steatosis by increasing nuclear SREBP-1C stability[107]. MALAT1 was also found to affect primary HSC proliferation, cell cycle, and activation by regulating RAS-associated C3 botulinum substrate 1 (Rac1) expression via the action of miR-101b as a ceRNA[106]. Accordingly, miR-101-3p downregulation was correlated with the presence of IR in preclinical models of NAFLD, and it actively contributed to disease progression through the activation of HSCs and hepatocyte proliferation. Indeed, insulin receptor haploinsufficient (InsR+/-) mice had reduced miR-101-3p expression compared to wild-type littermates, and miR-101-3p reduction prompted more severe histological liver damage. Interestingly, this phenomenon was exacerbated by exposure to a combination of insulin and fatty acids, resembling IR-NASH[108]. Thus, MALAT1- and IR-mediated downregulation of miR-101-3p may promote progressive NAFLD through HSC transdifferentiation and hepatocyte proliferation.
Highly upregulated in liver cancer (HULC)
The lncRNA HULC acts as an endogenous competitor for miR-2052, essential for softening the expression of MET, a tyrosine kinase that promotes tumorigenesis and tumor metastasis[109]. Elevated levels of HULC were reported in seven different human hepatocyte cell lines and HCC patients. Comparing HULC expression in 42 pairs of tumor tissues to non-cancerous ones, its upregulation resulted in MET activation, promoting cell growth and proliferation[110].
Increased HULC levels were also found in patients from Gene Expression Omnibus (GEO) datasets (GSE39791 and GSE76427) and The Cancer Genome Atlas (TCGA) cohort and were positively correlated with poor prognosis. miR-2052 expression, in turn, was dampened in HCC tissues and cells with increased HULC levels, suggesting a direct suppression of miR-2052 mediated by HULC[110]. Thus, the HULC/miR-2052/MET axis may be involved in the development of HCC and may be used as a novel prognostic biomarker and therapeutic target for HCC.
Nuclear enriched abundant transcript 1 (NEAT1)
NEAT1 is a lncRNA, which plays a pivotal role in adipogenesis by mediating LDL oxidization, lipid uptake, and lipolysis. NEAT1 has recently been found enriched in the liver of HCC and NAFLD patients and is associated with steatosis in HepG2 cells. NEAT1 expression aggravated free fatty acid (FFA)-induced lipid accumulation in hepatocytes by regulating the c-Jun/SREBP1c axis by sponging miR-139-5p. Indeed, NEAT1 heightening is followed by acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) mRNA enhancement in both in vivo and in vitro models of NAFLD[111]. A preclinical study showed that the knockdown of NEAT1 in hepatocytes hampered lipid content and liver disease through the modulation of ACC and FAS expression. As proof of NEAT1 actionability, another study reported the potential therapeutic role of NEAT1 downregulation against liver damage by inhibiting the mTOR/S6K1 pathway[111].
Homeobox (HOX) transcript antisense RNA (HOTAIR)
lncRNA HOTAIR promotes liver fibrosis in the NAFLD setting and could be an interesting target to attenuate liver disease. The expression of HOTAIR was reported to be boosted in the livers of CCl4-treated mice and correlated with HSC activation. In this model, HOTAIR acted as a ceRNA for miR-148B and mediated the activation of the DNMT1/MEG3/p53 pathway. Moreover, the intensification of HOTAIR expression was induced by fatty acids and inhibited by phosphatase and tensin homolog expression gene (PTEN). As a consequence, an increase in TG accumulation was observed in HepG2 steatotic cells and high-fat diet (HFD) mice[112]. Interestingly, the knockdown of HOTAIR significantly inhibited the progression of NAFLD in in vitro models, through the mediation of miR-130b-3p/ROCK1 signaling. In mice, HOTAIR knockdown inhibited lipid accumulation in HFD mice, restoring normal levels of serum TG, AST, and ALT and regulating the expression of genes implicated in lipid metabolism[113].
H19
A comprehensive study evaluated the impact of lncRNA H19 dysregulation on HCC severity in four different human HCC patient cohorts, H19 knockout mice, and three different human hepatoma cell lines. In detail, H19 downregulation was reported in tumors resected from 32 HCC patients compared to non-tumorous tissues, and it was directly correlated with tumor growth and chemoresistance. These findings were further confirmed on patients from public datasets. Accordingly, H19 knockout mice showed enhanced tumor development and growth. Sorafenib/doxorubicin-resistant cell lines exhibited reduced H19 expression compared to their chemosensitive counterparts. Resistance to therapy was reverted after H19 overexpression, which abolished tumor overgrowth and sensitized cells to treatment. Therefore, H19 may suppress carcinogenesis, tumor enlargement, and chemoresistance, thus constituting a promising agent to overcome chemoresistance in HCC therapy[114].
Circular RNAs
circRNAs also belong to the noncoding RNAs and are gaining increasing interest as biomarkers in different tumor types. circRNAs were initially described as highly stable scrambled exons resistant to endonucleases. Their structure consists predominantly of a circular loop RNA void of 5-cap and 3-tail containing miRNA response elements (MREs) and several conserved binding sites for miRNAs[115]. These peculiarities allow circRNAs to operate as “miRNA sponges”, able to impound miRNA transcripts, dysregulating their activity. Most circRNAs could interact with more than one miRNA and harbor one or more miRNA binding sites based on conserved seed sequence matches[116]. Therefore, circRNAs can effectively modulate miRNA transcription, further affecting the expression of downstream mRNA through a circRNA-miRNA-mRNA pathway. Various evidence indicates that some circRNAs are expressed aberrantly in HCC and regulate important processes involved in NAFLD worsening to HCC, such as lipogenesis, fibrosis, and cell proliferation[117].
Regulation of lipid metabolism by circRNAs
circRNA_0046367 and circRNA_0046366 were indicated as negative endogenous regulators of miR-34a, an inhibitor of peroxisome proliferator-activated receptor α (PPARα). Their altered expression was reported in NAFLD patients and steatotic cells[118]. Decreased expression of circRNA_0046367 was observed in liver tissues of NAFLD patients and in fat-laden HepG2 cells, while circRNA_0046366 was found reduced only in cell models. Here, circRNA_0046367 and circRNA_0046366 dysregulation prevented the complementary interaction with miR-34a, promoting the downregulation of PPARα and predisposing obese patients to IR and hepatic steatosis. Differently, when these two circRNAs were restored, PPARα expression increased, as well as its target gene carnitine palmitoyltransferase 2 (CPT2), acyl-CoA binding domain containing 3 (ACBD3), and solute carrier family 27A (SLC27A), thereby softening steatosis. These findings corroborate the hypothesis that abnormal regulation of circRNA_0046367 or circRNA_0046366/miR-34a/PPARα signaling may be a novel targetable epigenetic mechanism which triggers hepatic steatosis[118,119].
circRNA_021412, through the binding to its target, miR-1972, prevents Lipin 1 (LPIN1) downregulation. LPIN1 in hepatocytes has a dual role; it suppresses the lipogenic program by catalyzing the last step of TG synthesis and amplifies hepatic PGC-1α/PPARα signaling, which regulates fatty acid β-oxidation (FAO)[120]. Interestingly, Guo et al. reported, along with 357 other circRNAs possibly involved in NAFLD, the decrease of circRNA_021412 levels in HepG2 cells treated with fatty acids. circRNA_021412 downregulation attenuated its competitive inhibition to its target, resulting in dropped LPIN1 expression and steatogenesis. Therefore, dysregulation of the circRNA_021412/miR-1972/LPIN1 cascade may contribute to hepatic steatosis by impairing LPIN1 expression and disrupting the balance between lipogenesis and FAO[121].
circRNAs mediate the development of liver fibrosis
Recently, a study performed on thymosin beta 4 (Tβ4)-depleted LX-2, a human model of HSC, correlated the expression of circRNA-0067835 with liver fibrosis by the activation of Forkhead Box O3 (FOXO3a). Tβ4 has anti-inflammatory and antifibrotic activity, and its loss leads to severe fibrosis. Among 664 circRNAs found differentially expressed in Tβ4-depleted LX-2 cells, the increase circRNA_0067835 regulated FOXO3a expression, acting as a sponge for its downstream regulator, miR-155. FOXO3a has a pivotal role in integrating multiple pathways, such as PI3K/Akt, which may regulate HSC proliferation and trigger liver fibrosis. Notably, the activation of circRNA_0067835/miR-155/FOXO3a significantly fostered the expression of FOXO3a in the Tβ4-depleted LX-2 cells, highlighting circRNA-0067835 as a potential therapeutic target for patients with liver fibrosis, the main prognostic factor of HCC[122]. In addition, Chen et al. exploited a microarray analysis to investigate the comprehensive circRNA profile in HSCs exposed to radiation-induced liver fibrosis (RILF), which induces their activation. Intriguingly, 179 circRNAs were significantly upregulated and 630 circRNAs were downregulated in irradiated HSCs compared to untreated ones. Among the upregulated circRNAs, hsa_circ_0072765 can bind to miR-370, which suppresses the proliferation and activation of HSCs, attenuating hepatic fibrogenesis. This analysis also revealed the intensification of hsa_circ_0071410, which was significantly associated with RILF. Hsa_circ_0071410 sequestering miR-9-5p may inhibit HSC activation/proliferation and repress RILF[123,124].
Cell cycle regulation by circRNAs in HCC
Aberrant circRNAs in HCC patients and in in vitro models were reported to influence cell cycle progression contributing to tumor growth. A positive correlation between circ-ZEB1.33 upregulation and CDK6 activity was outlined in human HCC tissues and further confirmed in vitro. The cyclin D/CDK4/6 complex mediates the transition from the G1 to S phase of the cell cycle, promoting proliferation. Indeed, circ-ZEB1.33 increased levels avoid miR-200a-3p binding to its target, CDK6. Thus, cell proliferation was found to be prompted through the circ-ZEB1.33/miR-200a-3p/CDK6 axis[125]. Accordingly, hsa_circ_0016788 boosted tumorigenesis by regulating the CDK4 pathway through miR-486 sequestration. hsa_circ_0016788 was identified to be upregulated among a total of 1245 differently expressed circRNAs in HCC tissues compared to non-cancerous ones. As a consequence, hsa_circ_0016788 accelerates HCC growth via regulation of miR-486/CDK4[126]. Again, tumor growth was found to be mediated by exosome circ-deubiquitination (circ-DB) upregulation in HCC patients with higher body fat. In vivo and in vitro models confirmed that exo-circ-DB facilitates tumor enlargement through the suppression of miR-34a and the consequent activation of the USP7/Cyclin A2 signaling, an important regulator of cell cycle progression. Finally, other circRNAs, such as has_circ_0078710 and hsa_circ_0091581, were further correlated to HCC expansion, influencing cyclin E/CDK2, p53, and c-Myc activity, which are important modulators of the cell cycle[127].
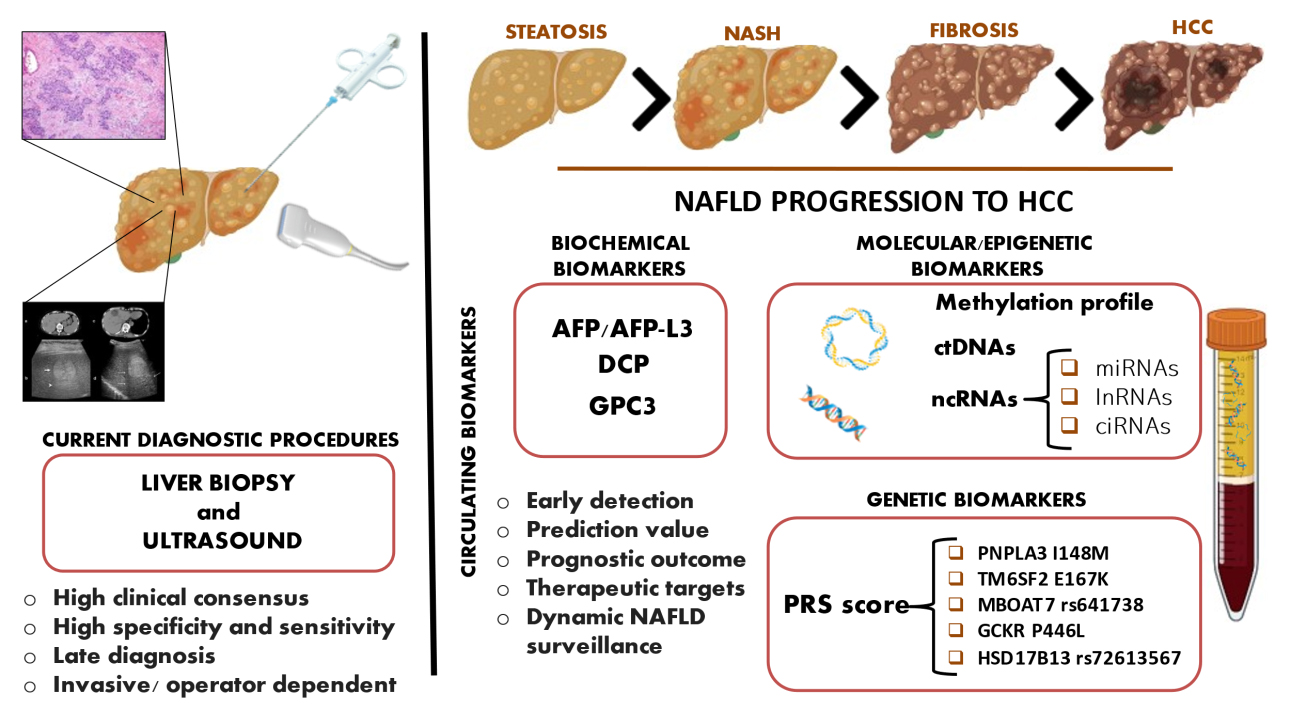
A schematic overview of old-style and new biomarkers is represented in Figure 1 and in Table 1.
Figure 1. Novel and old perspectives for HCC surveillance. Liver biopsy and ultrasound are currently the most used procedures to detect HCC, but these techniques harbor several limitations. Liquid biopsy offers a simple and non-invasive alternative and provides a wide range of potentially relevant molecules for tumor surveillance. Biochemical, epigenetic, and genetic biomarkers can be isolated from peripheral blood, and they are currently exploited in the HCC setting to ameliorate their detection. These markers can discriminate NAFLD stages, predict early HCC onset, and dynamically follow cancer evolution and metastasis.
Schematic overview of the main genetic, biochemical, molecular, and epigenetic biomarkers for NAFLD-HCC screening
| Biomarker | Hallmarks | Pros & Cons |
| Genetic | ||
| PRSs | Useful to stratify patients on the risk of disease progression to HCC High prognostic value | √ Non-invasive biomarkers √ Early HCC predictor √ Prognostic indicator of severe liver disease × Not clinically approved |
| Biochemical | ||
| AFP and AFP-L3 | Prognostic predictor of HCC Used to monitor HCC progression and metastasis Increased levels are found in HCC and during chronic liver disease | √ Non-invasive biomarker √ FDA approved √ Combined with abdominal US improve HCC detection × Not exclusive to HCC × Not suitable as a single biomarker for HCC screening × Low sensitivity × Not indicative of early HCC |
| DCP | Prognostic predictor of HCC Improves HCC surveillance after therapy High diagnostic efficacy for large and advanced tumors (high stage) | √ Non-invasive biomarker √ FDA approved √ Combined with AFP may complement US in the detection of early HCC √ Good in discriminating advanced tumor stages × Not suitable as a single biomarker for HCC screening × Not indicative of early HCC |
| Glypican 3 | Enables early HCC detection Novel target for HCC treatment Associated with poor HCC prognosis Indicative of tumor relapse after resection | √ Non-invasive biomarker √ Combined with AFP, gains in sensitivity and specificity √ Clinically actionable √ Ongoing clinical trials × Low sensitivity and poor detection rate in blood samples compared to tissue biopsies × Not suitable as a single biomarker for HCC screening |
| G-protein coupled receptor | Potential diagnostic biomarker Prognostic predictor of HCC - Beta2-AR is indicative of tumor recurrence after resection - G-protein signaling modulator 2 is enriched in HCC and correlates with tumor size - GPR50 expression is enhanced in HCC - Gα12 is involved in neoplastic transformation and is associated with a more aggressive phenotype | √ Non-invasive biomarkers √ Therapeutical target for the treatment of HCC × Not clinically approved × Beta2-AR role in HCC has not been thoroughly elucidated |
| Molecular | ||
| ctDNAs | Associated with poor HCC prognosis. Indicative for metastatic potential and tumor relapse | √ Non-invasive biomarkers √ Combined with AFP gains diagnostic value √ Predictive for ICB √ High sensitivity × Not exclusive to tumor condition × Not clinically approved |
| Noncoding RNAs | ||
| miRNAs | Involved in cancer diagnosis, prognosis, and therapeutic targets - miR-122 reduction is a direct inducer of NASH-associated HCC - miR-15/16 is downregulated in invasive HCC cell lines and in aggressive HCC - miR-221 and miR-101-3p overexpression stimulate cell growth and invasion | √ Non-invasive biomarkers √ Clinically actionable √ Dynamic HCC surveillance √ Available from multiple specimens (e.g. tumor tissues, peripheral blood, urine) × Research into miRNAs as biomarkers is still in its early stages × Lack of reproducibility and standardization of clinical protocols |
| lncRNAs | Correlated to HCC malignancy. Involved in the pathogenesis and development of HCC - MALAT1 is involved in cell proliferation, migration, invasion - HULC increased levels are correlated to a poor prognosis of HCC - NEAT1 expression aggravated FFA- induced lipid accumulation in hepatocytes - HOTAIR increasing promotes liver fibrosis in NAFLD setting - H19 is involved in tumor enlargement and chemoresistance | √ Non-invasive biomarkers √ Potential prognostic biomarkers for HCC √ Clinically actionable × lncRNA function not fully apprehended × Lack of reproducibility and standardization of clinical protocols |
| circRNAs | Useful to monitor NAFLD-HCC progression and indicative of early HCC - circRNA_0046367 and circRNA_0046366 altered expression may trigger hepatic steatosis - circRNA_021412 downregulation contributes to hepatic steatosis - circRNA-0067835 expression is correlated with liver fibrosis - hsa_circ_0072765 enhanced expression promotes liver fibrosis - hsa_circ_0071410 increased levels inhibit HSCs activation/proliferation and repress RILF - circ-ZEB1.33 is positively correlated with cell proliferation - hsa_circ_0016788 accelerates HCC growth - has_circ_0078710 and hsa_circ_0091581 are correlated to HCC expansion | √ Non-invasive biomarkers √ Clinically actionable × CircRNAs function is not fully clarified |
CONCLUDING REMARKS
Since about 25%-30% of the global population is affected by NAFLD, with an increasing trend in the future, NAFLD-driven HCC will become a huge socioeconomic burden in the next years[4]. HCC has an extremely poor survival rate, and this is partially due to late cancer diagnosis[128]. Currently, liver biopsy remains the gold standard procedure for the diagnosis of HCC, but it is not specific for early cancer detection and not suitable for mass screening. Indeed, it is an invasive procedure, potentially dangerous for the patient, and not feasible in children, elderly subjects, or patients with severe cirrhosis. Thus, there is an urgent need to implement surveillance programs focusing on new markers that can overcome biopsy[6]. New biomarkers and target molecules would allow a wide and non-invasive screening of the at-risk HCC population and the possibility of targeting specific dysregulated pathways during treatment. To date, there are several candidate molecules, some of which are already used in clinics to monitor the development of HCC. Well-consolidated molecular markers such as AFP and DCP have improved HCC surveillance, but several weak points have been pinpointed in discriminating HCC stage and worsening[45]. Furthermore, these biochemical markers cannot be used alone in a clinical setting due to their poor reliability and validity. Newly discovered genetic signatures, such as at-risk genetic variants combined in PRSs, epigenetic modifications, ctDNAs, and ncRNAs, are promising biomarkers with the potential to bridge the gap between the clinical requirements and the current needs[10,129]. In this review, we highlight the ability of these tools to discriminate the early stages of HCC, prevent the escalation of malignancy, and predict NAFLD evolution to HCC. Some of these new molecules, such as GCP3 and several ncRNAs, also show encouraging clinical applicability, being possible novel therapeutic targets against NAFLD-HCC[130]. The successful strategy to counteract the HCC burden may be the establishment of new diagnostic and clinical algorithms, integrating the use of “old” consolidated biomarkers with PRSs and the evaluation of ctDNA and different ncRNAs. This approach may lead to the early identification of HCC, the possibility to determine personalized therapeutic approaches based on the specific genetic/epigenetic/biochemical landscape, and consequently treat patients with targeted agents.
DECLARATIONS
Authors’ contributionsConceptualized the study and critically revised the manuscript: Meroni M, Dongiovanni P
Write the first version of the manuscript: Piciotti R, Longo M
Participate to the revision of the manuscript: Agresta A, Paolini E, Cespiati A
All Authors have read and agreed to the published version of the manuscript.
Availability of data and materialsNot applicable.
Financial support and sponsorshipThe study was supported by Italian Ministry of Health (Ricerca Corrente 2022, Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico, Milano) and Ricerca Finalizzata Ministero della Salute GR-2019-12370172.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
1. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73-84.
2. Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70.
4. Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2021;18:223-38.
5. Mittal S, El-Serag HB, Sada YH, et al. Hepatocellular carcinoma in the absence of cirrhosis in united states veterans is associated with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2016;14:124-31.e1.
6. Dongiovanni P, Meroni M, Longo M, Fargion S, Fracanzani AL. Genetics, immunity and nutrition boost the switching from NASH to HCC. Biomedicines 2021;9:1524.
7. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6.
8. Zhang H, Zhang W, Jiang L, Chen Y. Recent advances in systemic therapy for hepatocellular carcinoma. Biomark Res 2022;10:3.
9. Tabacelia D, Stroescu C, Dumitru R et al. New approach for hepatocellular carcinoma treatment. J Med Life 2022;15:138-43.
10. Meroni M, Longo M, Tria G, Dongiovanni P. Genetics is of the essence to face NAFLD. Biomedicines 2021;9:1359.
11. Valenti L, Dongiovanni P. Mutant PNPLA3 I148M protein as pharmacological target for liver disease. Hepatology 2017;66:1026-8.
12. Bruschi FV, Claudel T, Tardelli M, et al. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 2017;65:1875-90.
13. Dongiovanni P, Donati B, Fares R, et al. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol 2013;19:6969-78.
14. Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352-6.
15. Dongiovanni P, Petta S, Maglio C, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 2015;61:506-14.
16. Liu YL, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun 2014;5:4309.
17. Grandone A, Cozzolino D, Marzuillo P, et al. TM6SF2 Glu167Lys polymorphism is associated with low levels of LDL-cholesterol and increased liver injury in obese children. Pediatr Obes 2016;11:115-9.
18. Goffredo M, Caprio S, Feldstein AE, et al. Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: a multiethnic study. Hepatology 2016;63:117-25.
19. Mancina RM, Sentinelli F, Incani M, et al. Transmembrane-6 superfamily member 2 (TM6SF2) E167K variant increases susceptibility to hepatic steatosis in obese children. Dig Liver Dis 2016;48:100-1.
20. Donati B, Dongiovanni P, Romeo S, et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep 2017;7:4492.
21. Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology 2016;150:1219-1230.e6.
22. Luukkonen PK, Zhou Y, Hyötyläinen T, et al. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J Hepatol 2016;65:1263-5.
23. Meroni M, Longo M, Fracanzani AL, Dongiovanni P. MBOAT7 down-regulation by genetic and environmental factors predisposes to MAFLD. EBioMedicine 2020;57:102866.
24. Meroni M, Dongiovanni P, Longo M, et al. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine 2020;52:102658.
25. Yang J, Trépo E, Nahon P, et al. A 17-beta-hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatology 2019;70:231-40.
26. Ki Kim S, Ueda Y, Hatano E, et al. TERT promoter mutations and chromosome 8p loss are characteristic of nonalcoholic fatty liver disease-related hepatocellular carcinoma. Int J Cancer 2016;139:2512-8.
27. Buch S, Innes H, Lutz PL, et al. Genetic variation in TERT modifies the risk of hepatocellular carcinoma in alcohol-related cirrhosis: results from a genome-wide case-control study . Gut 2022;gutjnl-2022-327196.
28. Donati B, Pietrelli A, Pingitore P, et al. Telomerase reverse transcriptase germline mutations and hepatocellular carcinoma in patients with nonalcoholic fatty liver disease. Cancer Med 2017;6:1930-40.
29. Petersen KF, Dufour S, Hariri A, et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med 2010;362:1082-9.
30. Pelusi S, Baselli G, Pietrelli A, et al. Rare pathogenic variants predispose to hepatocellular carcinoma in nonalcoholic fatty liver disease. Sci Rep 2019;9:3682.
31. Meroni M, Longo M, Paolini E, et al. The rs599839 A>G variant disentangles cardiovascular risk and hepatocellular carcinoma in NAFLD patients. Cancers (Basel) 2021;13:1783.
32. Dongiovanni P, Meroni M, Petta S, et al. Neurotensin up-regulation is associated with advanced fibrosis and hepatocellular carcinoma in patients with MAFLD. Biochim Biophys Acta Mol Cell Biol Lipids 2020;1865:158765.
33. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med 2020;12:44.
34. Dongiovanni P, Stender S, Pietrelli A, et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med 2018;283:356-70.
35. Bianco C, Jamialahmadi O, Pelusi S, et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J Hepatol 2021;74:775-82.
36. Di Costanzo A, Pacifico L, Chiesa C, et al. Genetic and metabolic predictors of hepatic fat content in a cohort of Italian children with obesity. Pediatr Res 2019;85:671-7.
37. Suomela E, Oikonen M, Pitkänen N, et al. Childhood predictors of adult fatty liver. The cardiovascular risk in young Finns Study. J Hepatol 2016;65:784-90.
38. Xu M, Li Y, Zhang S, Wang X, Shen J, Zhang S. Interaction of TM6SF2 E167K and PNPLA3 I148M variants in NAFLD in northeast China. Ann Hepatol 2019;18:456-60.
39. Koo BK, Joo SK, Kim D, et al. Development and validation of a scoring system, based on genetic and clinical factors, to determine risk of steatohepatitis in Asian patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2020;18:2592-2599.e10.
40. Gellert-Kristensen H, Richardson TG, Davey Smith G, Nordestgaard BG, Tybjaerg-Hansen A, Stender S. Combined effect of PNPLA3, TM6SF2, and HSD17B13 variants on risk of cirrhosis and hepatocellular carcinoma in the general population. Hepatology 2020;72:845-56.
41. Longo M, Meroni M, Paolini E, et al. TM6SF2/PNPLA3/MBOAT7 Loss-of-function genetic variants impact on nafld development and progression both in patients and in in vitro models. Cell Mol Gastroenterol Hepatol 2022;13:759-88.
42. Hwang A, Shi C, Zhu E, et al. Supervised learning reveals circulating biomarker levels diagnostic of hepatocellular carcinoma in a clinically relevant model of non-alcoholic steatohepatitis; An OAD to NASH. PLoS One 2018;13:e0198937.
43. Lim TS, Rhee H, Kim GM, et al. Alpha-fetoprotein, des-gamma-carboxy prothrombin, and modified recist response as predictors of survival after transarterial radioembolization for hepatocellular carcinoma. J Vasc Interv Radiol 2019;30:1194-1200.e1.
44. Rojas Á, Sánchez-torrijos Y, Gil-gómez A, et al. Performance of different biomarkers for the management of hepatocellular carcinoma. HR 2018;4:31.
45. Pan Y, Chen H, Yu J. Biomarkers in hepatocellular carcinoma: current status and future perspectives. Biomedicines 2020;8:576.
46. Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the american association for the study of liver diseases. Hepatology 2018;68:723-50.
47. Di Bisceglie AM, Sterling RK, Chung RT, et al. ; HALT-C Trial Group. Serum alpha-fetoprotein levels in patients with advanced hepatitis C: results from the HALT-C trial. J Hepatol 2005;43:434-41.
48. Yang JD, Dai J, Singal AG, et al. Improved performance of serum alpha-fetoprotein for hepatocellular carcinoma diagnosis in HCV cirrhosis with normal alanine transaminase. Cancer Epidemiol Biomarkers Prev 2017;26:1085-92.
49. Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182-236.
50. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358-80.
51. Lok AS, Sterling RK, Everhart JE, et al. HALT-C Trial Group. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 2010;138:493-502.
52. Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology 2018;154:1706-1718.e1.
53. Zhou JM, Wang T, Zhang KH. AFP-L3 for the diagnosis of early hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 2021;100:e27673.
54. Nakamura S, Nouso K, Sakaguchi K, et al. Sensitivity and specificity of des-gamma-carboxy prothrombin for diagnosis of patients with hepatocellular carcinomas varies according to tumor size. Am J Gastroenterol 2006;101:2038-43.
55. Lee S, Rhim H, Kim YS, Kang TW, Song KD. Post-ablation des-gamma-carboxy prothrombin level predicts prognosis in hepatitis B-related hepatocellular carcinoma. Liver Int 2016;36:580-7.
56. Best J, Bechmann LP, Sowa JP, et al. GALAD score detects early hepatocellular carcinoma in an international cohort of patients with nonalcoholic steatohepatitis. Clin Gastroenterol Hepatol 2020;18:728-735.e4.
57. Zheng X, Liu X, Lei Y, Wang G, Liu M. Glypican-3: a novel and promising target for the treatment of hepatocellular carcinoma. Front Oncol 2022;12:824208.
58. Huang TS, Shyu YC, Turner R, Chen HY, Chen PJ. Diagnostic performance of alpha-fetoprotein, lens culinaris agglutinin-reactive alpha-fetoprotein, des-gamma carboxyprothrombin, and glypican-3 for the detection of hepatocellular carcinoma: a systematic review and meta-analysis protocol. Syst Rev 2013;2:37.
59. Liu X, Wang SK, Zhang K, et al. Expression of glypican 3 enriches hepatocellular carcinoma development-related genes and associates with carcinogenesis in cirrhotic livers. Carcinogenesis 2015;36:232-42.
60. Sun B, Huang Z, Wang B, et al. Significance of glypican-3 (GPC3) expression in hepatocellular cancer diagnosis. Med Sci Monit 2017;23:850-5.
61. Abou-Alfa GK, Puig O, Daniele B, et al. Randomized phase II placebo controlled study of codrituzumab in previously treated patients with advanced hepatocellular carcinoma. J Hepatol 2016;65:289-95.
62. Liu X, Gao F, Jiang L, et al. 32A9, a novel human antibody for designing an immunotoxin and CAR-T cells against glypican-3 in hepatocellular carcinoma. J Transl Med 2020;18:295.
63. Wu X, Luo H, Shi B, et al. Combined antitumor effects of sorafenib and GPC3-CAR T cells in mouse models of hepatocellular carcinoma. Mol Ther 2019;27:1483-94.
64. Chen D, Xing W, Hong J, et al. The beta2-adrenergic receptor is a potential prognostic biomarker for human hepatocellular carcinoma after curative resection. Ann Surg Oncol 2012;19:3556-65.
65. He XQ, Zhang YF, Yu JJ, et al. High expression of G-protein signaling modulator 2 in hepatocellular carcinoma facilitates tumor growth and metastasis by activating the PI3K/AKT signaling pathway. Tumour Biol 2017;39:1010428317695971.
66. Saha SK, Choi HY, Yang GM, et al. GPR50 promotes hepatocellular carcinoma progression via the notch signaling pathway through direct interaction with ADAM17. Mol Ther Oncolytics 2020;17:332-49.
68. Lombardi R, Piciotti R, Dongiovanni P, Meroni M, Fargion S, Fracanzani AL. PD-1/PD-L1 immuno-mediated therapy in NAFLD: advantages and obstacles in the treatment of advanced disease. Int J Mol Sci 2022;23:2707.
69. Gok Yavuz B, Hasanov E, Lee SS, et al. Current landscape and future directions of biomarkers for immunotherapy in hepatocellular carcinoma. J Hepatocell Carcinoma 2021;8:1195-207.
70. Pinter M, Jain RK, Duda DG. The current landscape of immune checkpoint blockade in hepatocellular carcinoma: a review. JAMA Oncol 2021;7:113-23.
71. Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med 1996;183:2533-40.
72. Sangro B, Melero I, Wadhawan S, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol 2020;73:1460-9.
73. Duffy AG, Ulahannan SV, Makorova-Rusher O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol 2017;66:545-51.
74. Feun LG, Li YY, Wu C, et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer 2019;125:3603-14.
75. Felden J, Garcia-Lezana T, Schulze K, Losic B, Villanueva A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut 2020;69:2025-34.
76. Dongiovanni P, Meroni M, Longo M, Fargion S, Fracanzani AL. miRNA signature in NAFLD: a turning point for a non-invasive diagnosis. Int J Mol Sci 2018;19:3966.
77. Pirola CJ, Gianotti TF, Burgueño AL, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2013;62:1356-63.
78. Kuramoto J, Arai E, Tian Y, et al. Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis 2017;38:261-70.
79. Tian Y, Wong VW, Wong GL, et al. Histone Deacetylase HDAC8 promotes insulin resistance and β-catenin activation in NAFLD-associated hepatocellular carcinoma. Cancer Res 2015;75:4803-16.
80. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84.
81. Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet 2016;12:e1006162.
82. Chan KC, Lai PB, Mok TS, et al. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem 2008;54:1528-36.
83. Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24.
84. Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: a broad overview. Crit Rev Oncol Hematol 2020;155:103109.
85. Cao SQ, Zheng H, Sun BC, et al. Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J Gastroenterol 2019;25:5283-99.
86. Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol 2017;67:603-18.
87. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer 2020;19:77.
88. Stroun M, Lyautey J, Lederrey C, Olson-sand A, Anker P. About the possible origin and mechanism of circulating DNA. Clinica Chimica Acta 2001;313:139-42.
89. Mezzalira S, De Mattia E, Guardascione M, Dalle Fratte C, Cecchin E, Toffoli G. Circulating-free DNA analysis in hepatocellular carcinoma: a promising strategy to improve patients' management and therapy outcomes. Int J Mol Sci 2019;20:5498.
90. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer 2019;18:114.
91. Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155-61.
92. Huang Z, Hua D, Hu Y, et al. Quantitation of plasma circulating DNA using quantitative PCR for the detection of hepatocellular carcinoma. Pathol Oncol Res 2012;18:271-6.
93. Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer 2020;1:873-81.
94. Rizzo A, Brandi G. Biochemical predictors of response to immune checkpoint inhibitors in unresectable hepatocellular carcinoma. Cancer Treat Res Commun 2021;27:100328.
95. Hsu C, Lu S, Abbas A, et al. Longitudinal and personalized detection of circulating tumor DNA (ctDNA) for monitoring efficacy of atezolizumab plus bevacizumab in patients with unresectable hepatocellular carcinoma (HCC). JCO 2020;38:3531-3531.
96. Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598-608.
97. Harding JJ, Nandakumar S, Armenia J, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res 2019;25:2116-26.
98. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013;12:847-65.
99. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004.
100. Fang Z, Dou G, Wang L. MicroRNAs in the pathogenesis of nonalcoholic fatty liver disease. Int J Biol Sci 2021;17:1851-63.
101. Lou G, Liu Y, Wu S, et al. The p53/miR-34a/SIRT1 positive feedback loop in quercetin-induced apoptosis. Cell Physiol Biochem 2015;35:2192-202.
102. Oura K, Morishita A, Masaki T. Molecular and functional roles of micrornas in the progression of hepatocellular carcinoma-a review. Int J Mol Sci 2020;21:8362.
103. IIott NE, Heward JA, Roux B, et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 2014;5:3979.
104. Huang R, Duan X, Fan J, Li G, Wang B. Role of noncoding RNA in development of nonalcoholic fatty liver disease. Biomed Res Int 2019;2019:8690592.
105. Leti F, Legendre C, Still CD, et al. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl Res 2017;190:25-39.e21.
106. Yu F, Lu Z, Cai J, et al. MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle 2015;14:3885-96.
107. Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep 2016;6:22640.
108. Meroni M, Longo M, Erconi V, et al. mir-101-3p downregulation promotes fibrogenesis by facilitating hepatic stellate cell transdifferentiation during insulin resistance. Nutrients 2019;11:2597.
109. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007;132:330-42.
110. Zhang H, Liao Z, Liu F, et al. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging (Albany NY) 2019;11:9111-27.
111. Jin SS, Lin CJ, Lin XF, Zheng JZ, Guan HQ. Silencing lncRNA NEAT1 reduces nonalcoholic fatty liver fat deposition by regulating the miR-139-5p/c-Jun/SREBP-1c pathway. Ann Hepatol 2022;27:100584.
112. Li W, Chen X, Lin M, Huang D. Up-regulated HOTAIR induced by fatty acids inhibits PTEN expression and increases triglycerides accumulation in HepG2 cells. Food & Nutr Res 2017;61:1412794.
113. Guo B, Cheng Y, Yao L, et al. LncRNA HOTAIR regulates the lipid accumulation in non-alcoholic fatty liver disease via miR-130b-3p/ROCK1 axis. Cell Signal 2022;90:110190.
114. Schultheiss CS, Laggai S, Czepukojc B, et al. The long non-coding RNA H19 suppresses carcinogenesis and chemoresistance in hepatocellular carcinoma. Cell Stress 2017;1:37-54.
116. Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci 2019;20:3926.
117. Zhang Y, Wang Y. Circular RNAs in hepatocellular carcinoma: emerging functions to clinical significances. Front Oncol 2021;11:667428.
118. Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 Prevents hepatoxicity of lipid peroxidation: an inhibitory role against hepatic steatosis. Oxid Med Cell Longev 2017;2017:3960197.
119. Guo XY, Sun F, Chen JN, Wang YQ, Pan Q, Fan JG. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol 2018;24:323-37.
120. Finck BN, Gropler MC, Chen Z, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab 2006;4:199-210.
121. Guo XY, He CX, Wang YQ, et al. Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis. Biomed Res Int 2017;2017:5936171.
122. Zhu L, Ren T, Zhu Z, et al. Thymosin-β4 mediates hepatic stellate cell activation by interfering with CircRNA-0067835/miR-155/FoxO3 signaling pathway. Cell Physiol Biochem 2018;51:1389-98.
123. Sun J, Zhang H, Li L, Yu L, Fu L. MicroRNA-9 limits hepatic fibrosis by suppressing the activation and proliferation of hepatic stellate cells by directly targeting MRP1/ABCC1. Oncol Rep 2017;37:1698-706.
124. Chen Y, Yuan B, Wu Z, Dong Y, Zhang L, Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of has_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene 2017;629:35-42.
125. Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int 2018;18:116.
126. Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol 2018;234:500-8.
127. Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019;38:2844-59.
128. Sim HW, Knox J. Hepatocellular carcinoma in the era of immunotherapy. Curr Probl Cancer 2018;42:40-8.
129. Han TS, Hur K, Cho HS, Ban HS. Epigenetic associations between LNCRNA/CIRCRNA and MIRNA in hepatocellular carcinoma. Cancers (Basel) 2020;12:2622.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Piciotti R, Longo M, Agresta A, Paolini E, Cespiati A, Meroni M, Dongiovanni P. Old-fashioned and newly discovered biomarkers: the future of NAFLD-related HCC screening and monitoring. Hepatoma Res 2022;8:37. http://dx.doi.org/10.20517/2394-5079.2022.46
AMA Style
Piciotti R, Longo M, Agresta A, Paolini E, Cespiati A, Meroni M, Dongiovanni P. Old-fashioned and newly discovered biomarkers: the future of NAFLD-related HCC screening and monitoring. Hepatoma Research. 2022; 8: 37. http://dx.doi.org/10.20517/2394-5079.2022.46
Chicago/Turabian Style
Piciotti, Roberto, Miriam Longo, Adele Agresta, Erika Paolini, Annalisa Cespiati, Marica Meroni, Paola Dongiovanni. 2022. "Old-fashioned and newly discovered biomarkers: the future of NAFLD-related HCC screening and monitoring" Hepatoma Research. 8: 37. http://dx.doi.org/10.20517/2394-5079.2022.46
ACS Style
Piciotti, R.; Longo M.; Agresta A.; Paolini E.; Cespiati A.; Meroni M.; Dongiovanni P. Old-fashioned and newly discovered biomarkers: the future of NAFLD-related HCC screening and monitoring. Hepatoma. Res. 2022, 8, 37. http://dx.doi.org/10.20517/2394-5079.2022.46
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 11 clicks
Cite This Article 11 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.