Association of human leukocyte antigen-DR-DQ-DP haplotypes with the risk of hepatitis B virus-related hepatocellular carcinoma
Abstract
Aim: Genetic polymorphisms of human leukocyte antigen (HLA) class II molecules are associated with chronic hepatitis B virus (HBV) infection. We aimed to investigate the impacts of HLA-II haplotypes on viral evolution and the risks of HBV-caused liver diseases.
Methods: HLA-DR-DQ-DP haplotypes were estimated in 1210 healthy controls, 296 HBV clearance subjects, 301 asymptomatic hepatitis B surface antigen carriers, 770 chronic hepatitis B patients, 443 HBV-related liver cirrhosis (LC) patients, and 1037 HBV-related hepatocellular carcinoma (HCC) patients. HBV mutations were determined by sequencing. The associations of HLA-DR-DQ-DP haplotypes with viral mutations and the risks of liver diseases were assessed by multivariate logistic regression.
Results: Compared to HBV-free subjects, the haplotypes CCAACG, CCGACG, TCAATA, and TCGATA were associated with decreased HCC risk, with an odds ratio (OR) [95% confidence interval (CI)] of 0.62 (0.40-0.95), 0.60 (0.39-0.92), 0.73 (0.54-0.98), and 0.58 (0.42-0.78), respectively. CCAACG, CCGACG, and TCAATA were significantly associated with decreased frequencies of the HCC-risk HBV mutations: preS1 deletion, APOBEC-signature HBV mutations in the core promoter and preS regions, A51C/T, G104C/T, and G146C/T. TCGATA and TTAACG were associated with increased LC risk, with an OR (95%CI) of 1.54 (1.03-2.30) and 2.23 (1.50-3.33), respectively. However, TCGATA and TTAACG were not consistently associated with the cirrhosis-risk HBV mutations.
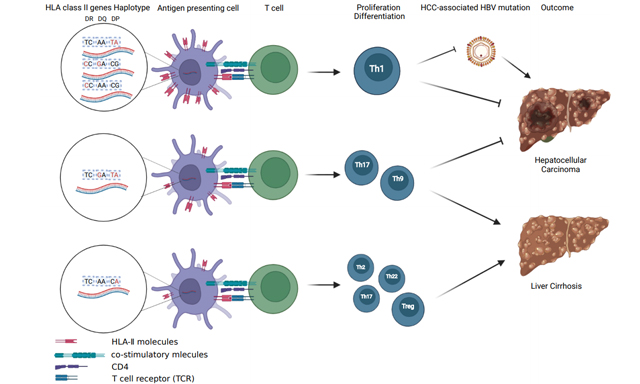
Conclusion: CCAACG, CCGACG, and TCAATA are inversely associated with HCC risk, possibly because they are involved in creating an immune microenvironment attenuating the generation of HCC-risk HBV mutations. TCGATA and TTAACG might predispose the polarity of immunity towards Th17 isotype related to LC.
Keywords
INTRODUCTION
Globally, primary liver cancer (PLC) is the sixth most common cancer and the third leading cause of cancer-related deaths, with an incidence-to-mortality ratio of nearly 1:1 (905,677 vs. 830,180 in 2020)[1]. China contributes to more than half of global PLC cases. The age-standardized mortality rate of PLC is higher in the middle-aged (40-64 years) population than in the elderly (≥ 65 years) population, in contrast to the other major cancers in China[2]. Hepatocellular carcinoma (HCC) accounts for 94.6% of PLC in eastern China. Although immunization of infants against hepatitis B virus (HBV) has reduced their risk of developing HCC, 87.5% of HCC is attributed to chronic HBV infection in eastern China[3,4]. There are approximately 94 million subjects chronically infected with HBV in mainland China[5]. We speculated that 32% male and 9% female subjects with chronic HBV infection might die of HCC before the average life (75 years old) in mainland China, based on the epidemiological data in Taiwan[6]. The key issue to decrease HCC-caused premature death is to identify the HBV-infected subjects who are more likely to develop HCC or postoperative relapse, which is essential for specific prophylaxis of this deadly malignancy.
HBV-induced hepatocarcinogenesis approximately takes 40-50 years. During this process, HBV experiences evolution via accumulating mutations in its genome, especially in the core promoter and the preS regions[7,8]. HBV is more apt to mutating than other DNA viruses due to the lack of proofreading function of the HBV genome during the reverse transcription process of pregenomic RNA, leading to a mutation rate of 2.2 × 10-5 substitutions/site/month in viral replication[9]. On the other hand, inflammatory molecules such as interleukin (IL)-6 trans-activate the expression of apoprotein B mRNA-editing enzyme catalytic polypeptides (APOBEC) 3B and inhibit the expression of uracil DNA glycosylasev, resulting in an increase in HCC-related viral mutations[10,11]. These HBV mutations may change viral replication, pathogenicity, and viral epitope that affect immune response, thus promoting the development of HCC[12-15]. Human leukocyte antigen (HLA) is mainly composed of HLA class I and class II molecules. HLA class I molecules may play a role in eliminating HBV-infected hepatocytes by presenting HBV peptides to CD8+ cytotoxic T lymphocytes. HLA class II molecules, which consist of three isotypes termed HLA-DR, HLA-DQ, and HLA-DP, can bind and present exogenous antigens to CD4+ T cells, thus initiating immune response. Several genome-wide association studies have shown that single nucleotide polymorphisms (SNPs) affecting the expression of HLA-II are associated with chronic HBV infection and HBV-related acute-on-chronic liver failure[16-18]. Our previous studies demonstrated that the genetic polymorphisms of HLA-DP, HLA-DQ, and HLA-DR are closely associated with chronic HBV infection, and the progression of liver diseases especially HBV-related HCC[19-21]. Several haplotypes of HLA class II genes were significantly associated with the response to an HBV vaccine[22]. However, few studies mentioned the association of HLA haplotypes with the risks of HBV-induced liver diseases. Here, we carried out an epidemiological study to investigate the association of haplotypes composed of several important HLA class II functional SNPs with the outcome of chronic HBV infection.
METHODS
Study subjects
This case-control study enrolled healthy controls, HBV natural clearance subjects, asymptomatic hepatitis B surface antigen (HBsAg) carriers (ASCs), chronic hepatitis B (CHB) patients, liver cirrhosis (LC) patients, and HBV-infected subjects with HCC. Healthy controls were recruited in subjects who received routine physical examination at the first affiliated hospital (Shanghai, China) of this university between September 2009 and August 2017. All healthy controls had no history of HBV and/or HCV infection and other liver diseases. HBV natural clearance subjects were defined as those who were seronegative for HBsAg and HBV DNA but seropositive for antibodies to both HBsAg and hepatitis B virus core antigen without HBV vaccination history. HBV natural clearance subjects and ASCs were recruited from our community-based epidemiological survey for HBV-infected individuals in Shanghai between September 2009 and August 2017. The patients with CHB, LC, and HCC were recruited from the first affiliated hospital between September 2009 and September 2017. The diagnostic criteria of ASCs, CHB, LC, and HCC were described in our previous studies[7,8]. At enrollment, we excluded CHB patients whose medical records showed a history of having received antivirals such as interferon, lamivudine, or adefovir or immunosuppressive treatments.
HLA class II genes genetic polymorphisms selection, genotyping, and haplotypes estimation
The haplotypes of HLA were estimated using tag SNPs. Six tag SNPs were involved in this study. rs3135338 (6400 bp upstream of HLA-DRA) and rs477515 (12 kb upstream of HLA-DRB1) were selected because they have been associated with chronic HBV infection and the risk of HBV-HCC[19]; rs2856718 and rs9275319 (located at the intergenic region between HLA-DQA2 and HLA-DQB1 region) were selected because they have been associated with the risk of chronic HBV infection and HCC in Chinese, Korean, and Japanese populations[20,23-26]; and rs3077 [located in the 3’ untranslated region (UTR) of HLA-DPA1] and rs9277535 (in the 3’ UTR of HLA-DPB1) were selected because they have been associated with the risk of chronic HBV infection and HCC[16,21,23]. Genomic DNA samples were extracted from peripheral lymphocytes using the blood genomic DNA extraction system (Tiangen, Beijing, China). HLA SNPs were genotyped using fluorescent-probe real-time quantitative PCR as previously described[19-21]. The sequences of primers and probes and PCR programs are shown in Supplementary Table 1. Hardy-Weinberg equilibrium for these six SNPs above was examined online (http://ihg.gsf.de/ihg/snps.html). The rates of successful genotyping calls were 98.2%-98.8% for these six SNPs.
HBV genotyping and viral mutation analysis
Extraction and quantification of HBV DNA and HBV genotyping was performed via multiplex and nested multiplex PCR according to our previously established methods[5]. The HBV core promoter region (nt1613-nt1849) and preS region (nt2848-nt155) were amplified and HBV mutations were determined by nested PCR as previously described[7,8]. The total numbers of APOBEC-signature viral mutations (C>T + C>G in TCW motif and G>A + G>C in WGA motif) in the HBV genome were calculated as previously described[10,11]. The APOBEC-signature mutation detected at ≥ 1 was defined as positive.
Statistical analysis
The “haplo.stats” package in R software (version 4.0.4) was used for assessing the association of HLA-DR-DQ-DP haplotype and viral mutations and disease status. The “haplo.score” function was used to estimate the frequency of each haplotype between different groups. The “haplo.glm” function was used for constructing the regression model of a trait on haplotype effects[27]. Haplotypes with frequencies higher than 2.5% in each group were selected, and the most frequent haplotype was chosen as the reference. Here, we chose TCAACG as the wild-type haplotype.
Differences in continuous variables such as age and serum HBV DNA load between two or more groups were evaluated by Student’s t-test and analysis of variance (ANOVA). Difference in categorical variables such as gander were evaluated by χ2 test. For the associations of haplotypes with the occurrence of HCC, clearance, persistence, and mutations of HBV, a logistic regression model was employed to calculate the odds ratio (OR) and 95% confidence interval (CI) in comparison to a common reference haplotype, adjusting for age and gender. All statistical tests were two-sided and performed using R statistical software (The R Foundation for Statistical Computing, Wien, Austria) and SPSS21.0 software (Statistical Product and Service Solutions, Chicago, USA). A P value of < 0.05 was considered statistically significant.
RESULTS
Characteristics of study subjects enrolled in the genotyping for HLA class II genes and haplotypes estimation
In total, 1210 healthy controls, 296 HBV natural clearance subjects, 301 ASCs, 770 CHB patients, 443 LC patients, and 1037 HCC patients were enrolled in this study. Sex proportion was not matched among healthy controls, HCC-free HBV-infected subjects, and HCC patients. Healthy controls were older than HCC-free HBV-infected subjects and HCC patients [Table 1]. Therefore, the OR values were all adjusted for gender and age. Haplotype was identified by the order of rs3135338, rs477515, rs2856718, rs9275319, rs3077, and rs9277535. The haplotypes with the frequencies > 2.5% (TCAACG, CCAACG, CCGACG, TCAACA, TCAATA, TCGACG, TCGATA, and TTAACG) were selected in the analyses.
The characteristics of study subjects enrolled in HLA class II genes haplotypes analysis
| Characteristic | Healthy controls (n = 1210) | HBV natural clearance subjects (n = 296) | HCC-free HBV-infected subjects (n = 1514) | HBV-infected subjects with HCC (n = 1037) | P value | ||
| ASCs (n = 301) | CHB (n = 770) | LC (n = 443) | |||||
| Male (%) | 870 (71.90) | 166 (56.08) | 177 (58.80) | 468 (60.78) | 325 (73.36) | 869 (83.80) | < 0.01a,b,c |
| Age, years, mean ± SD | 53.06 ± 17.49 | 58.54 ± 11.76 | 45.31 ± 10.73 | 53.63 ± 12.68 | 52.08 ± 11.02 | 52.30 ± 11.27 | < 0.01a, 0.11b, < 0.01c |
| HBV genotype (%) | |||||||
| B | - | - | 89 (33.21) | 90 (23.38) | 53 (21.20) | 102 (15.38) | < 0.01b |
| C | - | - | 179 (66.79) | 295 (76.62) | 197 (78.80) | 561 (84.62) | |
| HBV DNA load, log10 copies/mL, mean ± SD | - | - | 3.45 ± 1.38 | 3.79 ± 1.70 | 3.05 ± 2.09 | 3.11 ± 1.86 | 0.01b |
| ALT mean ± SD | 21.94 ± 14.72 | 25.45 ± 25.2 | 26.29 ± 17.53 | 92.73 ± 310.79 | 89.52 ± 151.31 | 72.08 ± 168.33 | < 0.01a,c, 0.16b |
Associations of HLA class II haplotypes with the risk of HCC
Compared to HBV-free subjects (healthy controls and HBV natural clearance subjects), the haplotypes CCAACG, CCGACG, TCAATA, and TCGATA were significantly associated with a decreased risk of HCC. In the genotype B HBV-infected subjects, TCAATA and TCGATA were associated with a decreased risk of HCC. In the genotype C HBV-infected subjects, TCGATA was associated with a decreased risk of HCC [Table 2]. Compared to healthy controls, CCAACG, TCAATA, and TCGATA were significantly associated with a decreased risk of HCC. The same was true for the genotype C HBV-infected subjects. Compared to the HBV natural clearance subjects, CCGACG was also associated with a decreased risk of HCC. This result was repeated in the genotype C HBV-infected subjects [Supplementary Table 2]. Compared to the HBV-infected subjects without HCC, CCAACG was associated with a decreased risk of HCC, while TCAACA was associated with a decreased risk of HCC in the genotype B HBV-infected subjects [Table 3].
Associations of HLA class II genes haplotypes with the risk of HCC
| HBV genotype | Haplotype | HBV-free subjects (healthy controls + HBV natural clearance subjects) (%) | HBV-HCC patients (%) | HBV-HCC patients vs. HBV-free subjects | |
| AOR (95%CI) | P value | ||||
| Subtotal | TCAACG | 19.02 | 27.60 | 1 | |
| CCAACG | 3.80 | 3.02 | 0.62 (0.40-0.95) | 0.03 | |
| CCGACG | 3.55 | 3.44 | 0.60 (0.39-0.92) | 0.02 | |
| TCAACA | 5.65 | 5.98 | 0.79 (0.57-1.09) | 0.15 | |
| TCAATA | 7.51 | 6.93 | 0.73 (0.54-0.98) | 0.04 | |
| TCGACG | 7.58 | 10.13 | 1.05 (0.77-1.41) | 0.77 | |
| TCGATA | 7.12 | 6.21 | 0.58 (0.42-0.78) | < 0.01 | |
| TTAACG | 5.03 | 4.03 | 0.85 (0.58-1.24) | 0.40 | |
| Genotype B | TCAACG | 19.02 | 33.14 | 1 | |
| CCAACG | 3.80 | 5.45 | 1.28 (0.80-2.07) | 0.73 | |
| CCGACG | 3.55 | 5.62 | 0.63 (0.34-1.18) | 0.51 | |
| TCAACA | 5.65 | 1.85 | 0.80 (0.51-1.24) | 0.06 | |
| TCAATA | 7.51 | 4.90 | 0.48 (0.29-0.80) | 0.02 | |
| TCGACG | 7.58 | 9.94 | 1.01 (0.67-1.52) | 0.75 | |
| TCGATA | 7.12 | 3.38 | 0.62 (0.40-0.96) | 0.01 | |
| TTAACG | 5.03 | 0.86 | 0.76 (0.45-1.28) | 0.06 | |
| Genotype C | TCAACG | 19.02 | 27.86 | 1 | |
| CCAACG | 3.80 | 3.45 | 0.82 (0.54-1.23) | 0.10 | |
| CCGACG | 3.55 | 3.01 | 0.75 (0.51-1.10) | 0.06 | |
| TCAACA | 5.65 | 5.20 | 0.74 (0.52-1.04) | 0.13 | |
| TCAATA | 7.51 | 7.18 | 0.80 (0.60-1.07) | 0.08 | |
| TCGACG | 7.58 | 10.02 | 1.06 (0.80-1.41) | 0.88 | |
| TCGATA | 7.12 | 5.88 | 0.61 (0.45-0.82) | < 0.01 | |
| TTAACG | 5.03 | 4.44 | 0.96 (0.69-1.34) | 0.83 | |
Associations of HLA class II genes haplotypes with HBV-infected subjects with HCC and HBV-infected subjects without HCC
| Genotype | Haplotype | HBV-infected subjects without HCC (%) | HBV-infected subjects with HCC (%) | HBV-infected subjects with HCC vs. HBV-infected subjects without HCC | |
| AOR (95%CI) | P value | ||||
| Subtotal | TCAACG | 25.61 | 27.60 | 1 | |
| CCAACG | 3.91 | 3.02 | 0.65 (0.44-0.97) | 0.04 | |
| CCGACG | 3.36 | 3.44 | 0.95 (0.64-1.40) | 0.79 | |
| TCAACA | 5.62 | 5.98 | 0.93 (0.67-1.28) | 0.63 | |
| TCAATA | 6.78 | 6.93 | 1.01 (0.75-1.35) | 0.96 | |
| TCGACG | 9.67 | 10.13 | 0.88 (0.67-1.15) | 0.36 | |
| TCGATA | 6.05 | 6.21 | 0.93 (0.69-1.24) | 0.62 | |
| TTAACG | 4.77 | 4.03 | 0.77 (0.56-1.06) | 0.10 | |
| Genotype B | TCAACG | 27.47 | 33.14 | 1 | |
| CCAACG | 7.22 | 5.45 | 0.61 (0.25-1.50) | 0.28 | |
| CCGACG | 3.21 | 5.62 | 1.51 (0.49-4.64) | 0.47 | |
| TCAACA | 6.30 | 1.85 | 0.29 (0.10-0.87) | 0.03 | |
| TCAATA | 4.71 | 4.90 | 0.62 (0.23-1.68) | 0.35 | |
| TCGACG | 10.05 | 9.94 | 1.07 (0.50-2.29) | 0.86 | |
| TCGATA | 6.79 | 3.38 | 0.44 (0.18-1.06) | 0.07 | |
| TTAACG | 4.21 | 0.86 | 0.29 (0.08-1.01) | 0.05 | |
| Genotype C | TCAACG | 24.78 | 27.86 | 1 | |
| CCAACG | 7.22 | 3.45 | 0.76 (0.43-1.34) | 0.34 | |
| CCGACG | 3.33 | 3.01 | 0.67 (0.39-1.13) | 0.13 | |
| TCAACA | 4.51 | 5.20 | 1.01 (0.59-1.73) | 0.75 | |
| TCAATA | 7.28 | 7.18 | 0.84 (0.55-1.26) | 0.38 | |
| TCGACG | 9.79 | 10.02 | 0.78 (0.53-1.14) | 0.30 | |
| TCGATA | 5.75 | 5.88 | 0.85 (0.55-1.32) | 0.55 | |
| TTAACG | 5.16 | 4.44 | 0.77 (0.50-1.20) | 0.24 | |
Associations of HLA class II haplotypes with the risks of chronic HBV infection, CHB, and LC
CCGACG was identified to be associated with a decreased risk of chronic HBV infection, compared to the HBV natural clearance subjects [Supplementary Table 3]. The Associations of HLA class II genes haplotypes with the risks of CHB and LC are shown in Table 4. Compared to ASCs, TCAACA and TTAACG were associated with an increased risk of CHB and LC. These effects were solely evident in the genotype C HBV-infected subjects. In addition, TCGACG was also associated with an increased risk of CHB and LC in the genotype C HBV-infected subjects. TCGATA and TTAACG were significantly associated with an increased risk of LC, compared to ASCs and CHB patients. Furthermore, TTAACG was significantly associated with an increased risk of LC in genotype C or B HBV-infected subjects [Table 5].
Associations of HLA class II genes haplotypes with the risk of CHB and LC
| Genotype | Haplotype | ASCs (%) | CHB and LC (%) | CHB and LC vs. ASCs | |
| AOR (95%CI) | P value | ||||
| Subtotal | TCAACG | 28.77 | 23.32 | 1 | |
| CCAACG | 4.68 | 4.20 | 0.93 (0.51-1.68) | 0.92 | |
| CCGACG | 5.21 | 2.96 | 0.75 (0.42-1.34) | 0.41 | |
| TCAACA | 2.40 | 6.17 | 2.43 (1.16-5.11) | 0.03 | |
| TCAATA | 8.05 | 6.36 | 1.01 (0.63-1.63) | 0.87 | |
| TCGACG | 8.06 | 9.78 | 1.46 (0.91-2.35) | 0.13 | |
| TCGATA | 5.29 | 5.86 | 1.14 (0.70-1.86) | 0.54 | |
| TTAACG | 3.73 | 5.69 | 1.77 (1.01-3.07) | 0.04 | |
| Genotype B | TCAACG | 26.98 | 24.44 | 1 | |
| CCAACG | 9.19 | 7.47 | 1.23 (0.44-3.47) | 0.66 | |
| CCGACG | 2.44 | 3.08 | - | ||
| TCAACA | 5.91 | 8.33 | 1.47 (0.45-4.74) | 0.62 | |
| TCAATA | 6.11 | 4.56 | 1.06 (0.32-3.44) | 0.93 | |
| TCGACG | 11.84 | 7.33 | 0.66 (0.25-1.71) | 0.17 | |
| TCGATA | 9.33 | 6.91 | 0.93 (0.39-2.22) | 0.95 | |
| TTAACG | 5.28 | 5.70 | 0.79 (0.28-2.17) | 0.63 | |
| Genotype C | TCAACG | 27.18 | 20.21 | 1 | |
| CCAACG | 3.99 | 3.21 | 0.77 (0.31-1.91) | 0.70 | |
| CCGACG | 5.67 | 3.10 | 0.65 (0.32-1.32) | 0.32 | |
| TCAACA | 1.55 | 4.98 | 3.84 (1.06-13.85) | 0.04 | |
| TCAATA | 9.76 | 6.78 | 1.00 (0.53-1.85) | 0.91 | |
| TCGACG | 6.47 | 11.20 | 2.63 (1.32-5.22) | < 0.01 | |
| TCGATA | 4.41 | 5.91 | 1.55 (0.71-3.40) | 0.23 | |
| TTAACG | 2.98 | 6.52 | 4.16 (1.60-10.81) | < 0.01 | |
Associations of HLA class II genes haplotypes with the risk of LC
| Genotype | Haplotype | ASCs and CHB (%) | LC (%) | LC vs. ASCs and CHB | |
| AOR (95%CI) | P value | ||||
| Subtotal | TCAACG | 25.36 | 21.78 | 1 | |
| CCAACG | 4.36 | 4.53 | 1.20 (0.72-1.99) | 0.49 | |
| CCGACG | 3.37 | 3.38 | 1.35 (0.79-2.31) | 0.28 | |
| TCAACA | 5.00 | 6.45 | 1.41 (0.90-2.23) | 0.14 | |
| TCAATA | 6.99 | 5.55 | 0.92 (0.59-1.45) | 0.73 | |
| TCGACG | 8.86 | 10.55 | 1.39 (0.95-2.04) | 0.09 | |
| TCGATA | 5.30 | 7.21 | 1.54 (1.03-2.30) | 0.03 | |
| TTAACG | 4.48 | 7.54 | 2.23 (1.50-3.33) | < 0.01 | |
| Genotype B | TCAACG | 28.28 | 17.99 | 1 | |
| CCAACG | 8.10 | 7.19 | 1.49 (0.49-4.53) | 0.49 | |
| CCGACG | 2.39 | 3.46 | 2.18 (0.48-10.02) | 0.32 | |
| TCAACA | 7.90 | 6.23 | 0.85 (0.29-2.54) | 0.77 | |
| TCAATA | 5.34 | 2.49 | 0.59 (0.09-3.87) | 0.59 | |
| TCGACG | 9.52 | 8.79 | 1.61 (0.53-4.83) | 0.4 | |
| TCGATA | 8.54 | 6.42 | 1.05 (0.34–3.30) | 0.93 | |
| TTAACG | 3.21 | 11.89 | 3.87 (1.35-11.13) | 0.01 | |
| Genotype C | TCAACG | 21.62 | 21.53 | 1 | |
| CCAACG | 3.96 | 3.21 | 0.79 (0.29-2.12) | 0.64 | |
| CCGACG | 3.94 | 3.58 | 1.00 (0.46-2.18) | 0.99 | |
| TCAACA | 3.51 | 5.47 | 1.50 (0.68-3.33) | 0.31 | |
| TCAATA | 8.52 | 5.78 | 0.75 (0.39-1.43) | 0.38 | |
| TCGACG | 9.73 | 10.8 | 1.19 (0.68-2.07) | 0.71 | |
| TCGATA | 4.41 | 7.12 | 1.45 (0.78-2.73) | 0.24 | |
| TTAACG | 5.17 | 7.13 | 1.88 (1.05-3.35) | 0.03 | |
Associations of HLA class II genes haplotypes with LC-related and HCC-related HBV mutations
We successfully amplified and sequenced the core promoter and preS regions from 1311 (83.72%) and 1161 (74.14%) of all HBV-infected subjects, respectively (GenBank accession Nos. JX556943-JX559050, KC934199-KC934744, and KJ019219-KJ019299). Supplementary Table 4 and 5 show the associations of HBV mutations with the risk of HCC and LC after the adjustment for age, gender, and HBV DNA load. A1762T, G1764A, A1762T/G1764A, C9A, A30T, T48A, T53C, C75A, G104C, C108T, A134C, G146C, and A3215C were significantly associated with an increased risk of HCC in both genotype B and C HBV-infected subjects. Interestingly, APOBEC-signature mutations in the core promoter and the PreS regions, C1653T, T1674C, T1753C, A1846T, C6A, A51T, PreS1 deletion, PreS2 deletion, and PreS deletion were significantly associated with an increased risk of HCC solely in genotype C HBV-infected subjects. APOBEC-signature mutations in the core promoter and the PreS regions, T1753C, A1762T, G1764A, C1766T, A1762T/G1764A, T1768A, C6A, A3215C, and PreS1 deletion were significantly associated with an increased risk of LC in both genotype B and C HBV-infected subjects. C1653T, A1726C, G1809A, C9A, T48A, A51C/T, T2857C, and PreS2 deletion were significantly associated with an increased risk, whereas T1674C/G and C75A were associated with a decreased risk, of LC solely in genotype C HBV-infected subjects.
The associations of HLA class II haplotypes with the frequencies of these LC-related HBV mutations are shown in Supplementary Table 6. CCGACG was associated with an increased frequency of T1674C/G. TCGACG was associated with increased frequencies of C6A, C1653T, and C1766T and a decreased frequency of C75A. TCGATA was associated with an increased frequency of T1674C/G. TTAACG was associated with increased frequencies of T1674C/G and T48A.
The associations of HLA class II haplotypes with the frequencies of the HCC-related HBV mutations are shown in Table 6. CCAACG was associated with a decreased frequency of the preS1 deletion. CCGACG was associated with decreased frequencies of APOBEC-signature mutations in both regions, A51C/T, G104C/T, and G146C/T. TCAATA was associated with a decreased frequency of APOBEC-signature mutations in the core promoter region.
Significant associations of HLA class II genes haplotypes with frequencies of HCC-associated HBV mutations
| Haplotype | HCC-associated HBV mutation | AOR (95%CI) | P value |
| TCAACG | 1.00 | ||
| CCAACG | preS1 deletion | 0.34 (0.12-0.94) | 0.04 |
| CCGACG | APOBEC-signature mutations in the core promoter region (< 2 vs. ≥ 2) | 0.35 (0.13-0.96) | 0.04 |
| APOBEC-signature mutations in the preS region (< 3 vs. ≥ 3) | 0.35 (0.13-0.93) | 0.03 | |
| A51C/T | 0.46 (0.21-0.99) | 0.04 | |
| G104C/T | 0.45 (0.21-0.97) | 0.04 | |
| G146C/T | 0.45 (0.21-0.98) | 0.04 | |
| TCAATA | APOBEC-signature mutations in the core promoter region (< 2 vs. ≥ 2) | 0.49 (0.26-0.92) | 0.03 |
DISCUSSION
The outcomes of chronic HBV infection depend on two aspects: viral genotype and host immunity. Compared with genotype B HBV-infected subjects, genotype C was more apt to cause chronic infection[28]. HLA-II whose genes encode both the alpha and beta chains has been proven to be the major immune factors affecting the outcomes of HBV infection[16-26]. HLA-II molecules are normally expressed on antigen presenting cells including Kupffer cells, the resident macrophages in liver, and play key roles in the presentation of viral antigens to CD4+ T helper (Th) lymphocytes. Th1-mediated immune response facilitates HBV clearance and inhibits the development of HCC, while Th2-, Th22-, Th17-, and Treg-related inflammation may predispose HBV persistence, poor prognosis of HBV-related acute-on-chronic liver failure, and cirrhosis[29-31]. Genetic predisposition of the three genes HLA-DR, HLA-DQ, and HLA-DP affects the host immunity and inflammation, mostly via affecting their expression. The genotypes that tend to upregulate their expression upregulate their immune responses[19,32]. However, it remains a great challenge to know what kinds of expression patterns of HLA-DR, HLA-DQ, and HLA-DP contribute to the polarity towards Th1 or other Th isotypes.
In this study, we found that the haplotype CCAACG with a “mutation” at HLA-DR rs3135338 and “wild-types” at other five SNPs was significantly associated with a decreased risk of HCC, compared to HBV-free subjects and HBV-infected subjects without HCC, indicating the upregulation of HLA-DRA might predispose the Th1 response to reduce the risk of HCC. Similarly, TCAATA with two “mutations” at HLA-DP functional SNPs rs3077 and rs9277535 was significantly associated with a decreased risk of HCC compared to HBV-free subjects, indicating an increased transcription of HLA-DPA1 and HLA-DPB1 in normal liver might predispose the Th1 immune response too. Interestingly, TCAACA with one “mutation” at HLA-DP rs9277535 was associated with an increased risk of benign liver diseases (CHB + LC) compared to ASCs, possibly because HLA-DPB1 in normal liver predisposes to Th2-, Th22-, Th17-, and Treg-related inflammation. TTAACG with one “mutation” at rs477515 was associated with an increased risk of LC compared to ASCs and CHB patients, indicating that high expression of HLA-DRB1 alone might be sufficient to induce the polarity towards higher Th17/Treg ratio, which is related to HBV-caused liver fibrosis[19,31,33]. Interestingly, TCGATA with one “mutation” at HLA-DQ rs2856718 and two “mutations” at HLA-DP rs3077 and rs9277535 was significantly associated with a decreased risk of HCC and an increased risk of LC [Tables 2 and 5], possibly because the co-expression of HLA-DQ, HLA-DPA1, and HLA-DPB1 might contribute to the polarity towards higher Th9- and Th17-mediated immune responses in liver. Th17 cells differentiate from Th0 naïve T cells in response to transforming growth factor β1 and interleukin-6, which drive the development of liver fibrosis[34]. HBV-specific Th9 cells are decreased in HBV-related HCC patients[35]. Thus, the haplotypes of HLA-DR, HLA-DQ, and HLA-DP might predispose the polarity of immune response upon HBV infection and even the consequences of chronic HBV infection.
An insufficient and persistent antiviral immune reaction can contribute to the generation of the HBV mutations during HBV evolution. We hypothesize that different expression profiles of HLA-DR, HLA-DQ, and HLA-DP might affect the development of HCC or LC via influencing the generation of HCC- or LC-related HBV mutations. In this study, the haplotypes promoting HBV persistence were found to be associated with a higher prevalence of HCC-risk HBV mutations, indicating that the effect of the haplotypes on the development of HCC might be indirectly influenced by the generation of HCC-risk HBV mutations. The immune polarity regulated by the expression profile of HLA isotypes might maintain the proinflammatory microenvironment for the generation and immune selection of HCC-risk HBV mutations. APOBECs whose expressions are trans-activated by IL-6 and other inflammatory factors can cause APOBEC-signature mutations in the HBV genome[10]. APOBEC-signature mutations can promote the development of HCC[11]. Thus, APOBECs can bridge chronic hepatitis and HCC. However, the effect of the haplotypes with the development of LC is unlikely to be influenced by the generation of LC-risk HBV mutations. The haplotype TTAACG was associated with an increased risk of LC, but it was positively associated with T1674C/G and T48A, whereas T1674C/G was associated with a decreased risk of LC compared to ASCs and CHB patients [Supplementary Table 5]. Thus, the effect of TTAACG on the development of LC should be determined by other pathways such as Th17-related signaling pathways.
The haplotype analysis provides an important option to know the effects of one or more SNPs of the three HLA-II isotypes on the development of HBV-related diseases especially HCC. It also provides an excellent opportunity to know which one or two of the HLA-DR, HLA-DQ, and HLA-DP expressions contribute to the polarity towards Th1 or other T helper isotypes. However, our study has several limitations. First, cases and controls were not matched on age and gender. HCC is more frequent in males than in females[1,6]. Second, the sample size was small, especially for those with HBV-related LC and HBV natural clearance subjects, resulting in the uncertainty of some associations. Third, the sample size of the HBV-infected subjects whose HBV fragments have been both amplified and sequenced was small, resulting in loss of data concerning HBV mutations. Further studies with larger sample sizes are needed to validate our findings. Fourth, although we excluded people who received antiviral treatment, other potential confounders might also influence the results.
In summary, the present study suggests that the haplotypes CCAACG, CCGACG, TCAATA, and TCGATA at rs3135338, rs477515, rs2856718, rs9275319, rs3077, and rs9277535 are significantly associated with a decreased risk of HCC. These haplotypes are also associated with decreased frequencies of most HCC-risk HBV mutations including the APOBEC-signature mutations in the core promoter and preS regions of the HBV genome. These data provide epidemiological evidence regarding the effect of haplotypes of HLA class II alleles in chronic HBV infection, viral evolution, and HCC development, which help provide genetic clues for choosing proper prophylactic and therapeutic options for HBV-infected subjects.
DECLARATIONS
Authors’ contributionsStudy concept and design: Cao G
Patient enrollment: Yin J, Tan X, Tan X, Han X
Identification of HBV mutations: Chen Y, Deng Y, Li Z, Zhou X, Cai S, Pu R
Host genome analysis: Chen Y, Deng Y, Liu W
Data analysis: Chen Y, Lin J, Deng Y
Manuscript drafting: Chen Y, Deng Y
Extensive revision and interpretation: Cao G
Study supervision: Cao G
Availability of data and materialsAll data of this article will be shared with publication upon reasonable request.
Financial support and sponsorshipThis study was supported by the National Natural Science Foundation of China (91529305, 81520108021, and 81673250 to GC) and the National Key Basic Research Program of China (973 program) (2015CB554006 to GC).
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateAll study subjects were genetically unrelated ethnic Han Chinese and provided written informed consent. The study was performed in accordance with the 2000 Declaration of Helsinki and was approved by the ethics committee of Second Military Medical University.
Consent for publicationAll study subjects were genetically unrelated ethnic Han Chinese and provided written informed consent.
Copyright© The Author(s) 2022.
REFERENCES
1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209-49.
2. Jiang D, Zhang L, Liu W, et al. Trends in cancer mortality in China from 2004 to 2018: a nationwide longitudinal study. Cancer Commun (Lond) 2021;41:1024-36.
3. Yang F, Ma L, Yang Y, et al. Contribution of hepatitis B virus infection to the aggressiveness of primary liver cancer: a clinical epidemiological study in Eastern China. Front Oncol 2019;9:370.
4. Wang XY, Huang JM, Lu XM, et al. Changing risk factors for hepatocellular carcinoma in hyperendemic regions in the era of universal hepatitis B vaccination. Cancer Epidemiol 2020;67:101775.
5. Yin J, Zhang H, He Y, et al. Distribution and hepatocellular carcinoma-related viral properties of hepatitis B virus genotypes in Mainland China: a community-based study. Cancer Epidemiol Biomarkers Prev 2010;19:777-86.
6. Huang YT, Jen CL, Yang HI, et al. Lifetime risk and sex difference of hepatocellular carcinoma among patients with chronic hepatitis B and C. J Clin Oncol 2011;29:3643-50.
7. Yin J, Xie J, Liu S, et al. Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am J Gastroenterol 2011;106:81-92.
8. Yin J, Xie J, Zhang H, et al. Significant association of different preS mutations with hepatitis B-related cirrhosis or hepatocellular carcinoma. J Gastroenterol 2010;45:1063-71.
9. Lim SG, Cheng Y, Guindon S, et al. Viral quasi-species evolution during hepatitis Be antigen seroconversion. Gastroenterology 2007;133:951-8.
10. Liu W, Wu J, Yang F, et al. Genetic Polymorphisms predisposing the interleukin 6-induced APOBEC3B-UNG imbalance increase HCC risk via promoting the generation of APOBEC-signature HBV mutations. Clin Cancer Res 2019;25:5525-36.
11. Yin J, Chen X, Li N, et al. Compartmentalized evolution of hepatitis B virus contributes differently to the prognosis of hepatocellular carcinoma. Carcinogenesis 2021;42:461-70.
12. Liang YJ, Teng W, Chen CL, et al. Clinical implications of HBV PreS/S mutations and the effects of PreS2 deletion on mitochondria, liver fibrosis, and cancer development. Hepatology 2021;74:641-55.
13. Anugwom CM, Allaire M, Akbar SMF, et al. Hepatitis B-related hepatocellular carcinoma: surveillance strategy directed by immune-epidemiology. Hepatoma Res 2021;7:23.
14. Tai DI, Tai J. The role of genetic factors in HBV-related HCC: perspectives from local genetic backgrounds and clinical epidemiology. Hepatoma Res 2020;6:74.
15. Lin CL, Kao JH. Prevention of hepatitis B virus-related hepatocellular carcinoma. Hepatoma Res 2021;7:9.
16. Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009;41:591-5.
17. Hu Z, Liu Y, Zhai X, et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet 2013;45:1499-503.
18. Tan W, Xia J, Dan Y, et al. Genome-wide association study identifies HLA-DR variants conferring risk of HBV-related acute-on-chronic liver failure. Gut 2018;67:757-66.
19. Deng Y, Li P, Liu W, et al. The genetic polymorphism down-regulating HLA-DRB1 enhancer activity facilitates HBV persistence, evolution and hepatocarcinogenesis in the Chinese Han population. J Viral Hepat 2020;27:1150-61.
20. Ji X, Zhang Q, Li B, et al. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol 2014;28:201-9.
21. Zhang Q, Yin J, Zhang Y, et al. HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J Virol 2013;87:12176-86.
22. Nishida N, Sugiyama M, Sawai H, et al. Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology 2018;68:848-58.
23. Jiang DK, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 2013;45:72-5.
24. Jiang DK, Ma XP, Yu H, et al. Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology 2015;62:118-28.
25. Kim LH, Cheong HS, Namgoong S, et al. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci of STAT4 and HLA-DQ in a Korean population. Infect Genet Evol 2015;33:72-6.
26. Mbarek H, Ochi H, Urabe Y, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet 2011;20:3884-92.
27. Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425-34.
28. Zhang HW, Yin JH, Li YT, et al. Risk factors for acute hepatitis B and its progression to chronic hepatitis in Shanghai, China. Gut 2008;57:1713-20.
29. Gu Y, Chen L, Lian Y, et al. Serum HBV pregenomic RNA is correlated with Th1/Th2 immunity in treatment-naïve chronic hepatitis B patients. J Med Virol 2020;92:317-28.
30. Mo R, Wang P, Lai R, et al. Persistently elevated circulating Th22 reversely correlates with prognosis in HBV-related acute-on-chronic liver failure. J Gastroenterol Hepatol 2017;32:677-86.
31. Li X, Su Y, Hua X, et al. Levels of hepatic Th17 cells and regulatory T cells upregulated by hepatic stellate cells in advanced HBV-related liver fibrosis. J Transl Med 2017;15:75.
32. O'Brien TR, Kohaar I, Pfeiffer RM, et al. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 2011;12:428-33.
33. Li K, Liu H, Guo T. Th17/Treg imbalance is an indicator of liver cirrhosis process and a risk factor for HCC occurrence in HBV patients. Clin Res Hepatol Gastroenterol 2017;41:399-407.
34. Li N, Yamamoto G, Fuji H, Kisseleva T. Interleukin-17 in liver disease pathogenesis. Semin Liver Dis 2021;41:507-15.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Chen Y, Lin J, Deng Y, Liu W, Li Z, Zhou X, Cai S, Pu R, Yin J, Tan X, Zhao J, Han X, Cao G. Association of human leukocyte antigen-DR-DQ-DP haplotypes with the risk of hepatitis B virus-related hepatocellular carcinoma. Hepatoma Res 2022;8:8. http://dx.doi.org/10.20517/2394-5079.2021.133
AMA Style
Chen Y, Lin J, Deng Y, Liu W, Li Z, Zhou X, Cai S, Pu R, Yin J, Tan X, Zhao J, Han X, Cao G. Association of human leukocyte antigen-DR-DQ-DP haplotypes with the risk of hepatitis B virus-related hepatocellular carcinoma. Hepatoma Research. 2022; 8: 8. http://dx.doi.org/10.20517/2394-5079.2021.133
Chicago/Turabian Style
Chen, Yifan, Jiansheng Lin, Yang Deng, Wenbin Liu, Zishuai Li, Xinyu Zhou, Shiliang Cai, Rui Pu, Jianhua Yin, Xiaojie Tan, Jun Zhao, Xue Han, Guangwen Cao. 2022. "Association of human leukocyte antigen-DR-DQ-DP haplotypes with the risk of hepatitis B virus-related hepatocellular carcinoma" Hepatoma Research. 8: 8. http://dx.doi.org/10.20517/2394-5079.2021.133
ACS Style
Chen, Y.; Lin J.; Deng Y.; Liu W.; Li Z.; Zhou X.; Cai S.; Pu R.; Yin J.; Tan X.; Zhao J.; Han X.; Cao G. Association of human leukocyte antigen-DR-DQ-DP haplotypes with the risk of hepatitis B virus-related hepatocellular carcinoma. Hepatoma. Res. 2022, 8, 8. http://dx.doi.org/10.20517/2394-5079.2021.133
About This Article
Copyright
Data & Comments
Data

 Cite This Article 19 clicks
Cite This Article 19 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.