3D Organoid modelling of hepatoblast-like and mesenchymal-like hepatocellular carcinoma cell lines
Abstract
Aim: We wished to establish 3D organoid-like hepatocellular carcinoma (HCC) models from HCC cell lines.
Methods: Hep3B, Huh7, HepG2, SNU398, SNU449, and SNU475 cell lines were inoculated into Matrigel and grown up to 9 days in hepatocyte specific or standard RPMI media. Spheroid formation was followed by light microscopy. Matrigel scaffolds were immobilized and embedded in paraffin, and sections were subjected to H&E and immunohistochemical staining for different hepatobiliary biomarkers. Stained material was examined under light microscopy and micro photo were taken.
Results: Organoid-like structures were obtained successfully from all selected cell lines except mesenchymal-like SNU475 cells. Hep3B, Huh7, and HepG2 cell lines from hepatoblast-like sub-group formed compact 3D colonies and showed hepatocyte-like morphology and staining with different hepatocyte lineage markers as well as hepatobiliary progenitor markers. SNU398 and SNU449 cell lines from mesenchymal-like group formed irregular and loose 3D colonies that expressed vimentin homogeneously, but also several epithelial and hepatocyte lineage markers. The pattern of biomarker expression was unique for each cell line tested. Such features, not observed in tested monolayer cultures were confirmed with single-cell derived Hep3B cells.
Conclusion: We described experimental conditions to obtain organoid-like structures from five different HCC cell lines representing hepatoblast-like and mesenchymal-like subgroups. These models are useful as an alternative to monolayer cultures to study phenotypic features of HCC cells. Our detailed analysis of biomarker expression in five different organoid-like structures provide convincing evidence for highly specific phenotypic features of these cell lines although they share some common or subtype-restricted features also.
Keywords
INTRODUCTION
Established cell lines derived from tumors of human hepatocellular carcinoma (HCC) have been widely used as models for studying cancer biology including mechanisms of proliferation, invasion, and metastasis[1-5]. They are also useful to establish models for acquired resistance to anticancer compounds. Currently, different HCC cell lines are available worldwide and they represent a necessary source for basic and translational research on HCC[5]. Indeed, although several laboratories have reported the establishment of tumor organoids (or tumoroids) from primary HCC tissues, this practice is not yet widely available[6,7]. More importantly, access to primary liver tumors is quite limited due to the underlying liver disease as well as in vivo diagnostic techniques that relived in vitro techniques in recent years.
Moreover, most of the available HCC cell lines have been thoroughly characterized by multi-omic techniques in terms of their mutation status, genome-wide mRNA, and mi-RNA expression profiles as well as protein expression studies by proteomics[4,5]. However, there are some limitations of established HCC cell lines such as their capacity to represent the well-known heterogeneity of HCC tumors which can only be addressed by establishing additional cell lines. Another major limitation of available cell-lines is their traditional cultures on plastic as monolayers 2D cultures. This monolayer culture practice is far from representing three-dimensional growth of primary tumors.
Similar to primary tumors, HCC cell lines form several subgroups. Fuchs et al.[2] identified subgroups of epithelial-like and mesenchymal-like cells that are based on the expression of E-cadherin and vimentin, respectively. We refer to them as “well-differentiated” and “poorly differentiated” subgroups, respectively[3]. More recently, Caruso et al.[5] examined molecular characteristics by mutation analysis, transcriptome analysis, and proteomics, and identified three subgroups namely CL1, CL2, and CL3 also qualified as hepatoblast-like, mixed epithelial-mesenchymal, and mesenchymal-like, respectively. Here we tried to establish 3D cell culture methods to generate organoid-like structures from three hepatoblast-like and three mesenchymal-like HCC cell lines. Organoid-like structures have been obtained successfully from given cell lines and we compared their morphologies together with HCC marker protein expression patterns identified by immunocytochemistry. Our primary intention was to optimize organoid culture conditions with standard cell culture media in order to obtain organoid-like characteristics under simple conditions at affordable costs. After establishing organoid-type 3D cell culture conditions, we also optimized conditions for paraffin embedding, as well as histochemical and immunohistochemical techniques easily available in a clinical pathology laboratory setting. We also describe here morphological and protein marker expression features of six HCC cell lines representative of hepatocyte-like and mesenchymal-like subgroups.
METHODS
Cell lines
Hep3B, Huh7, and HepG2 cell lines from hepatoblast-like and SNU398, SNU449, and SNU475 cell lines from mesenchymal-like subgroups of HCC were tested for their ability to form organoid-like structures under 3D growth conditions. The origins and subgroups of cell lines have been described previously[1,3,5]. Single cell derived Hep3BTAp73β cells from parental Hep3B cell line[8] were also used for control experiments.
Monolayer cell culture
Following defrosting, cell lines were first cultivated as monolayers for at least three passages in RPMI (Cat. 11875168, Thermo Fisher Scientific, Waltham, MA, USA) containing non-essential amino acid mixture (1X) (Cat.10270106, Thermo Fisher Scientific), L-Glutamine (2 mM), penicillin-streptomycin (100 U and 100 µg, respectively) (Cat. 15140148, Thermo Fisher), and 10% (v/v) fetal calf serum (FBS) (Cat. 16000036, Thermo Fisher Scientific). Cells were routinely cultured in a humidified atmosphere at 37 °C and 5% CO2.
Optimization of Matrigel scaffold-based 3D organotypic culture system
We established organoid-like structures from HCC cell lines, based on previously published conditions described for 3D organoid (or tumoroid) culture of primary tumor samples[9,10]. To optimize cell number in 3D culture, cells were prepared in either “hepatocyte culture medium” or “RPMI medium” and seeded into 24-well plate. Hepatocyte culture medium complemented with epidermal growth factor (Corning Cat. 355056) was mixed with 5% charcoal-stripped serum (Gibco, Cat. 12676029) as described by Chua et al.[9] and Lee et al.[10]. The complete RPMI medium was used as described above for the monolayer cell culture.
After the third 2D cell culture passage, 4 × 104 or 8 × 104 cells in 60 µL cell culture medium mixed with
Preparation of paraffin blocks from HCC cell-line organoid-like spheroids
The 3D organoid-like spheroids formed on the 9th day were harvested using cold (+4 °C) PBS and fixed in 10% formalin (Fisher Chemical) for 30 min. Organoid-like spheroids were processed for paraffin sectioning using plasma thrombin clot preparation technique[11]. Hematoxylin-eosin (H&E) staining was performed using standard protocols on 5 µm paraffin sections. Histologic evaluation of H&E-stained sections of organoid-like spheroids samples was performed by a pathologist.
Histology and immunohistochemical staining
For immunochemical staining, 4-5 µm thick sections were prepared from paraffin-embedded tissues and were placed on electrostatic-charged slides (X-traTM, Surgipath Medical Industries, Richmond, Illinois, USA). All Sections were deparaffinized and dehydrated through a graded ethanol series using routine protocols. Immunohistochemistry study was performed usingmouse or rabbit proper monoclonal antibodies. Briefly, sections were incubated at 50 °C overnight, deparaffinized in xylol, and rehydrated through graded alcohol rinses. Antigen retrieval, blocking, and the incubation with monoclonal antibodies were performed automatically on Ventana Platform as a staining antibody. Primary antibodies to the following antigens were used: Ki67 [Dako (MIB-I), 1/100], E-cadherin [VENTANA (36), ready to use], vimentin [Ventana (U9), ready to use], CD44 [VENTANA (SP37), ready to use], smooth muscle actin [Cell mergue (1A4), Ventana, 1/300], Axl (C89E7) Rabbit mAb #8661 Cell Signaling, 1:75 delusion, catalog number: 8661S, beta-catenin [Cell marque (14), Ventana, ready to use], cytokeratin 7 [Ventana, ready to use], cytokeratin 18 [Epitomics (EP30), 1/100], cytokeratin 19 [CELL MARQUE (A52-B/A2.26), ready to use], cytokeratin 20 [VENTANA (SP33), ready to use], HEPpar1 [CELL MARQUE (OCH1E5), ready to use], alpha-fetoprotein [CELL MARQUE (POLYCLONAL), 1/100], Alpha1-antitrypsin [Dako (AAT) 1/30], glutamine-synthetase [Cell mergue (G5-6), Ventana, 1/200], and arginase [CELL MARQUE (SP156), ready to use]. Full list of primary antibodies is provided in Supplementary Table 1. The secondary antibody kit is used accordingly with the Ventana Platform. It used a positive control for each antibody accordingly, whereas primary mAb was omitted for negative control sections. For comparison of 3D organoids with monolayer cultures, Hep3B and SNU449 cell lines were grown as monolayers on sterilized microscope slides, fixed with cold ethanol for 5 min and subjected to immunohistochemical staining using a Ventana automatic staining device. The slides were analyzed under an Olympus BX61 microscope (Olympus Corporation, Tokyo, Japan).
Nuclear staining of Ki67, regardless of its intensity, was considered to be positive. The labelling-index was expressed as a percentage of positive nuclei[12]. Stained organoid like spheroids were analyzed and classified as negative, heterogeneously positive, or homogeneously positive.
RESULTS
We selected six HCC cell lines, three from hepatoblast-like and another three from mesenchymal-like subgroups[5] to establish 3D organoid-like models in order to develop an alternative model to monolayer culture that have major limitations. The mutational status of selected cell lines was given in Table 1.
| Hep3B | Huh7 | HepG2 | SNU398 | SNU449 | SNU475 |
| TP53e | TP53c | TERTb | Tp53a | TP53c | TP53c |
| AXIN1a | TERTb | ATMc | TERTb | ARID1Aa | TERTb |
| RPS6KA3e | FGF19f | CTNNB1d | TSC2e | CDKN2Ae | AXIN1e |
| RB1a | CCND1f | FAT4c | CTNNB1c | AKAP9c | FGF19f |
| KMT2Da | PREX2c | FLT4c | MTAPe | CCND1f | |
| PREX2c | NRASc | ZNF521c | ARID2c | AKAP9c | |
| PDE4DIPa | ZFHX3c | KEAP1a | |||
| G6PCc | PTENc | ACVR2Ac | |||
| HNF1Ac |
Matrigel was used as a scaffold. 1-2 × 104 cells were incubated in 50 µL Matrigel droplets in two different cell culture media, one based on the use of hepatocyte culture medium[9], the other standard RPMI medium. 3D cell cultures have been observed every other day until day 9. The experiments were stopped upon observation of large number of colonies under light microscopy [Supplementary Figure 1]. Colonies were fixed, paraffin embedded, 5 µm sections were cut and treated for histochemical and immunochemical staining. Best results were obtained with 2 × 104 starting cell number and 8-10 days of cell culture, independent of cell culture media [Supplementary Figure 2]. Therefore, follow up studies were carried out in standard RPMI cell culture conditions.
Organoid-like structures from hepatoblast-like cell lines
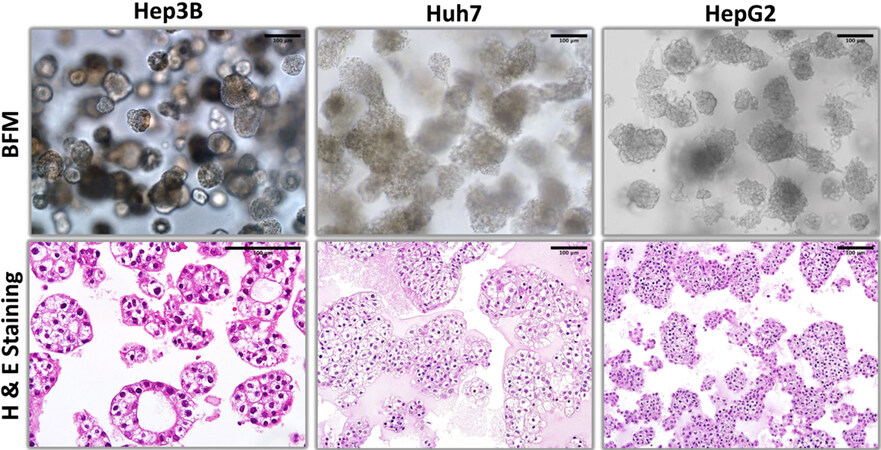
HCC-derived Hep3B and Huh7 cell lines and hepatoblastoma-derived HepG2 cell line from the “hepatoblast-like” subgroup were grown under 3D culture conditions and examined at day 9 [Figures 1-5]. These cells formed large grape-like colonies each composed of tens to hundreds of cells depending on the cell line [Figure 1]. Hep3B cells formed smaller colonies of two major types. Some of these colonies were adenoid-like acinar structures with noticeable lumen[13], while others were filled with regular shaped cells, as seen with H&E staining [Figure 1]. Huh7 cells formed larger colonies and formed large clear cells. In contrast, HepG2 colonies were formed with intensely staining small cells [Figure 1].
Figure 1. Bright field microscopy and hematoxylin-eosin staining patterns of organoid-like structures observed with hepatoblast-like hepatocellular carcinoma cell lines.
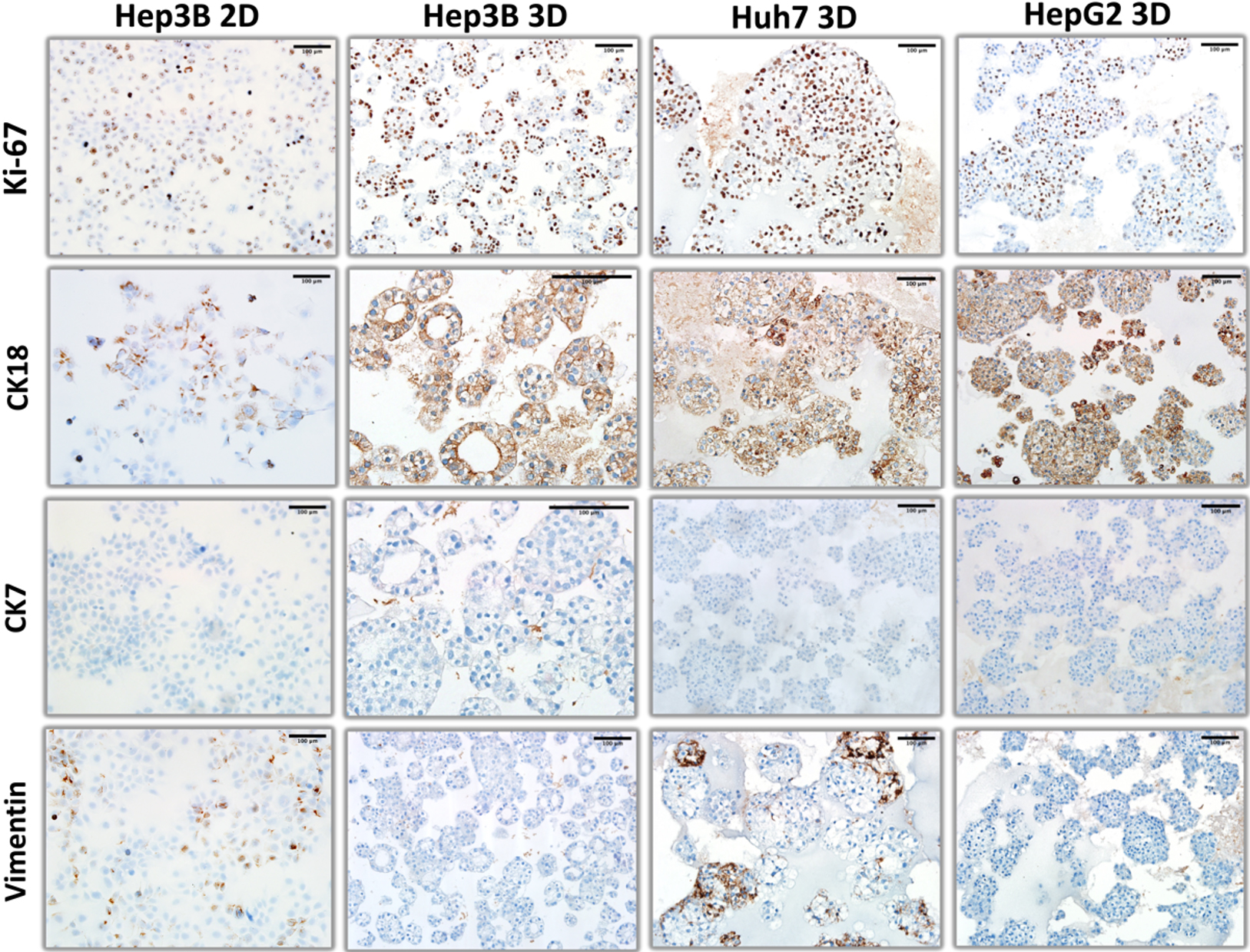
Figure 2. Comparative immunohistochemical analysis of Ki-67, CK18, CK7, and vimentin expression in hepatoblast-like cell lines. Hep3B cells were used to illustrate different patterns of expression of these proteins under 2D monolayer (Hep3B 2D) and 3D organoid-like (Hep3B 3D) culture conditions. For two other hepatoblast-like cell lines (Huh7 and HepG2) pictures represent immunostaining patterns under 3D organoid-like culture conditions.
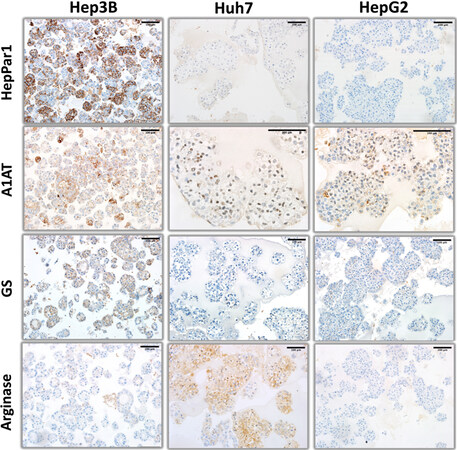
Figure 3. Hepatocyte lineage markers (HepPar1, alpha-1 antitrypsin, glutamine synthetase, and arginase) immunohistochemistry staining patterns of organoid-like structures observed with hepatoblast-like hepatocellular carcinoma cell lines.
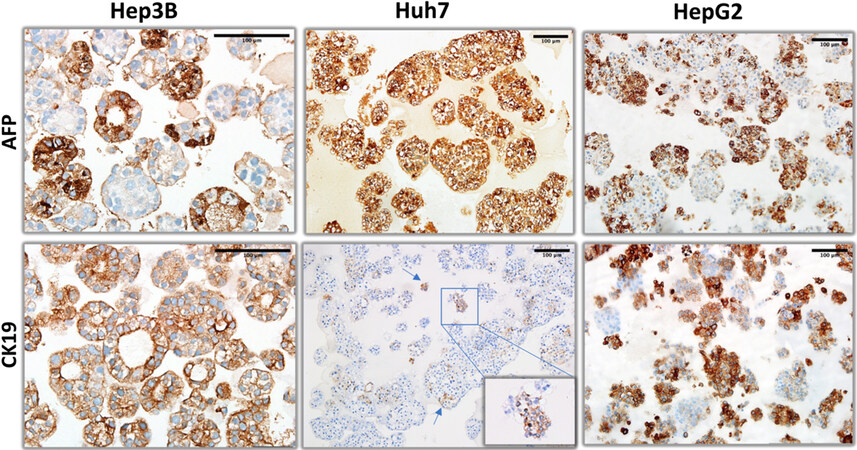
Figure 4. Liver progenitor lineage markers (alpha-fetoprotein and CK19) immunohistochemistry staining patterns of organoid-like structures observed with hepatoblast-like hepatocellular carcinoma cell lines.
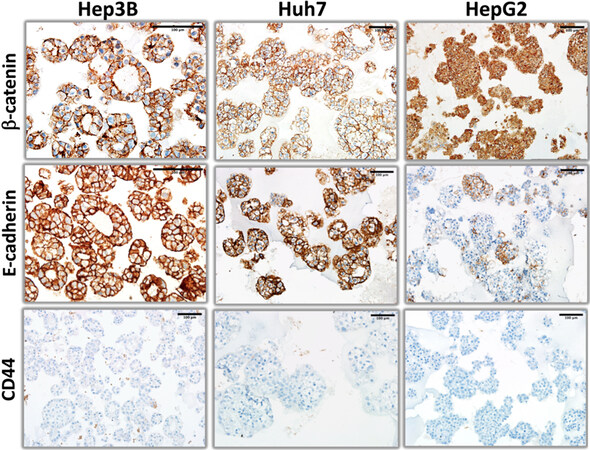
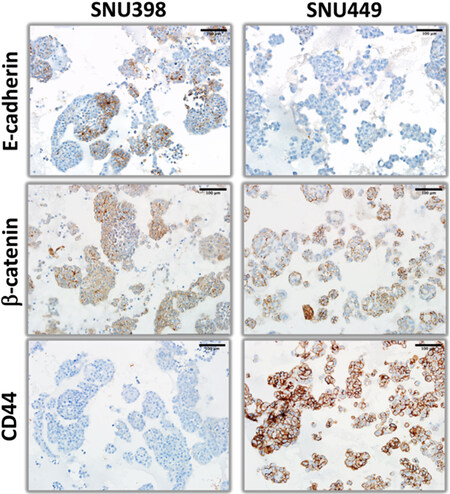
Figure 5. β-catenin, E-cadherin, and CD44 immunohistochemistry staining patterns of organoid-like structures observed with hepatoblast-like hepatocellular carcinoma cell lines.
The proliferation patterns of these cell lines, as tested by Ki-67 staining were clearly different from each other [Figure 2]. Hep3B cells were positive mostly at peripheral cells with about 50% positivity. HepG2 cells were also partly positive but proliferating cells were scattered among Ki-67-negative cells irregularly. In contrast, Huh7 cells displayed a highly proliferative morphology with nearly 100% of cells being Ki67-positive [Figure 2]. We also analyzed the expression of intermediate filaments including hepatocyte-associated cytokeratin 18 (CK18), biliary cell-marker CK7[14] and mesenchymal cell marker vimentin in these hepatoblast-like cell lines [Figure 2]. All three displayed positive staining for CK18, but negative staining for CK7 and vimentin as an indication of their hepatocyte-like phenotype. CK18 staining pattern in acinar-like Hep3B structures was intense at apical site near the lumen, as an indication of differentiation. In contrast, Huh7 and HepG2 showed diffuse cytoplasmic staining. vimentin was mostly negative with few positive colonies in only Huh7 cells [Figure 2].
We compared the organization and biomarker expression in HCC cells under 3D growth conditions with features under 2D monolayer culture. Hep3B was selected as a model for hepatoblast-like cells [Figure 2]. and SNU449 as representatives of mesenchymal-like HCC cell lines (see later in figure 7). Hep3B cells did not form any colonies under 2D conditions. There was no lumen formation. Cells were scattered with no major contact with each other. When stained for Ki-67, the ratio of positive cells did not show a striking difference between 3D and 2D growth conditions. For CK7 and CK18 expression there was no major difference between different growth conditions, cells being negative for CK7 and nearly fully positive for CK18. In contrast, Hep3B cells grown as monolayers showed higher positivity for vimentin, compared to their growth under 3D conditions. Another major difference between 3D and 2D cell structures was the subcellular localization of CK18 expression. Its expression was restricted mostly at the periphery of the cytoplasm in 3D organoids, whereas it displayed diffuse cytoplasmic and perinuclear staining in cells grown as monolayers (compare Hep2B 2D and Hep3B 3D panel photos in Figure 2).
The expression of hepatocyte markers was positive but showed heterogeneity in hepatoblast-like HCC cell lines. Hep3B, but not the other two cell lines displayed mostly strong, but regionally weak granular staining for HepPar1, a mitochondrial antigen associated with hepatocytes. Hep3B and HepG2 cells displayed colonies staining positively for A1AT (alpha-1 antitrypsin), a liver-specific protein secreted to the plasma. Huh7 cells were uniformly but weakly positive for A1AT with some unexpected nuclear staining. Hep3B cells were also positive for glutamine synthetase, but not for arginase. In contrast, Huh7 cells expressed arginase but not glutamine synthetase. HepG2 cells were negative for both markers [Figure 3].
Next, we explored the expression of hepatobiliary progenitor cell markers, AFP (alpha-fetoprotein) and CK19[14]. Hep3B and HepG2 cells formed AFP+ and AFP- cell colonies, whereas Huh7 cells were homogeneously positive. The pattern of CK19 staining was also different between cell lines. Hep3B cells were homogeneously positive, whereas HepG2 cells formed positive as well as negative colonies. Finally, CK19 staining of Huh7 cells were extremely rare and weakly positive [Figure 4]. Epithelial-mesenchymal transition defines the ability of epithelial cells to gain a mesenchymal morphology[15]. In order to test the ability of hepatoblast-like cells to undergo epithelial-mesenchymal transition under 3D growth conditions, we performed immunostaining for E-cadherin and β-catenin as epithelial markers, and CD44 as mesenchymal marker [Figure 5] in addition to vimentin presented in Figure 2. Both Hep3B and Huh7 displayed fully positive membrane-localized E-cadherin and β-catenin staining in all colonies tested. HepG2 cells displayed a few E-cadherin-positive colonies, β-catenin staining in HepG2 was strong and mostly diffuse rather than membrane-localized. Similarly, CD44 staining was negative in all three cell lines [Figure 5].
Organoid-like structures from mesenchymal-like cell lines
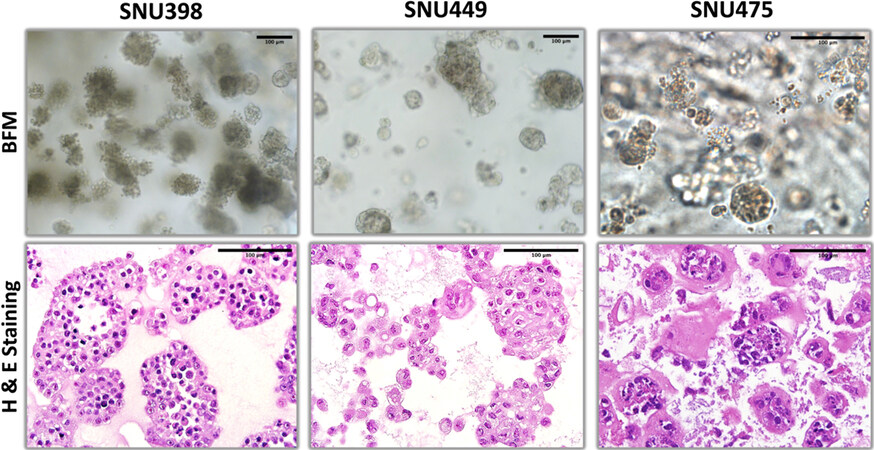
Three cell lines selected from the mesenchymal-like HCC subgroup were explored for organoid-like growth ability. SNU398 and SNU449 cell lines were able to create organized 3D structures [Figure 6], but SNU449 3D structures were less cohesive compared to 3D structures observed with hepatoblastoma-like cell lines [Figure 1]. SNU475, on the other hand, did not form organized structures under our experimental conditions. As seen by H&E staining, SNU475 cells formed only small and scattered cell groups (consequently, we did not further explore this cell line under 3D growth conditions). Of particular interest, lumen-forming structures associated with glandular epithelium were not observed in mesenchymal-like SNU398 and SNU449 cell lines. This was expected as they are classified as mesenchymal-like cell lines.
Figure 6. Bright field microscopy and hematoxylin-eosin staining patterns of organoid-like structures observed with mesenchymal-like hepatocellular carcinoma cell lines.
Ki67 immunohistochemical proliferative index of SN398 was about 40%, while SNU449 displayed a much lower proliferation index [Figure 7]. We observed strong and homogenous vimentin expression in these cell lines classified as mesenchymal-like cells. However, the expression of hepatocyte-associated cytoskeletal protein CK18 was negative in SNU398 as expected, but surprisingly positive in some 3D colonies of SNU449 cells. Interestingly, SNU449 cells but not SNU338 cells expressed biliary cell-associated cytoskeletal marker CK7, almost fully [Figure 7].
Figure 7. Comparative immunohistochemical analysis of Ki-67, CK18, CK7, and vimentin expression in mesenchymal-like cell lines. SNU449 cells were used to illustrate different patterns of expression of these proteins under 2D monolayer (SNU449 2D) and 3D organoid-like (SNU449 3D) culture conditions. For two SNU 398 pictures represent immunostaining patterns under 3D organoid-like culture conditions.
As stated before, we also compared SNU449 cells after growth under 2D monolayer (SNU449 2D) and 3D organoid-like (SNU449 3D) conditions [Figure 7]. SNU449 cells did not show colony-like organization as monolayer. The rates of positive staining for Ki67 and CK18 were similar under 3D and monolayer conditions. Subcellular organization of their expression was not selective under monolayer culture conditions displaying diffuse cytoplasmic and perinuclear patterns. Similar differences were also observed with CK7 staining.
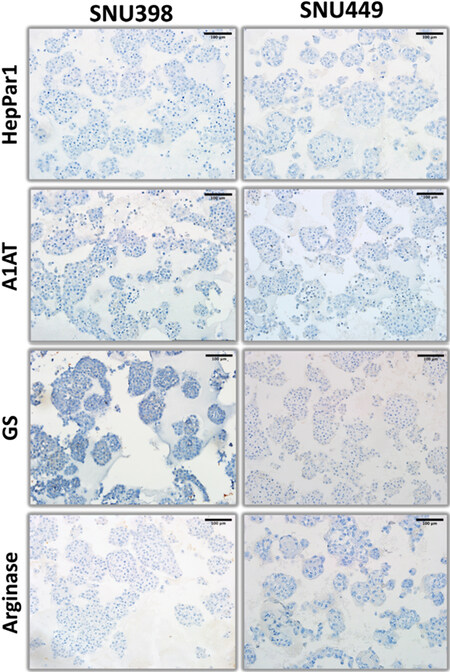
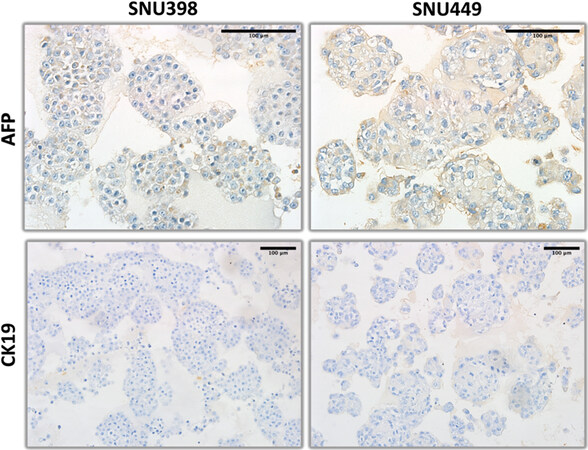
The expression of hepatocyte markers was negative in SNU398 and SNU449 [Figure 8]. In terms of progenitor markers, AFP-positive colonies were detected in both SNU398 and SNU449 3D structures, but CK19 expression was negative in both cell lines [Figure 9]. The study of the expression of epithelial-mesenchymal transition markers in these two cell lines indicated that SNU398, but not SNU449 cells were able to generate some epithelial-like E-cadherin positive colonies. The β-catenin was weakly, but uniformly positive in both cell lines. Finally, strong membrane staining for CD44 was observed in SNU449, but not in SNU398 cells [Figure 10].
Figure 8. Hepatocyte lineage markers (HepPar1, alpha-1 antitrypsin, glutamine synthetase, and arginase) immunohistochemistry staining patterns of organoid-like structures observed with mesenchymal-like hepatocellular carcinoma cell lines.
Figure 9. Liver progenitor lineage markers (alpha-fetoprotein and CK19) immunohistochemistry staining patterns of organoid-like structures observed with mesenchymal-like hepatocellular carcinoma cell lines.
Figure 10. β-catenin, E-cadherin, and CD44 immunohistochemistry staining patterns of organoid-like structures observed with mesenchymal-like hepatocellular carcinoma cell lines.
Our immunostaining data for five cell lines was summarized in Table 2.
Expression of biomarkers in organoid-like structures of hepatocellular carcinoma cell lines
| Cell Line | Hep3B | Huh7 | HepG2 | SNU398 | SNU449 |
| Subgroup | HB-like | HB-like | HB-like | MES-like | MES-like |
| Ki67 | 55% Pos. | 90% Pos. | 40% Pos. | 50% Pos. | 40% Pos. |
| HepPar1 | Het. Pos. | Neg. | Neg. | Neg. | Neg. |
| GS | Het. Pos. | Neg. | Neg. | Neg. | Neg. |
| Arginase | Neg. | Het. Pos. | Neg. | Neg. | Neg. |
| A1AT | Het. Pos. | Het. Pos.* | Het. Pos. | Neg. | Neg. |
| CK18 | Hom. Pos. | Hom. Pos. | Hom. Pos. | Neg. | Het. Pos. |
| CK7 | Neg. | Neg. | Neg. | Neg. | Pos. |
| CK19 | Hom. Pos. | Rare. Pos. | Het. Pos. | Neg. | Neg. |
| AFP | Het. Pos. | Hom. Pos. | Het. Pos. | Het. Pos. | Het. Pos. |
| E-cadherin | Hom. Pos. | Hom. Pos. | Het. Pos. | Het. Pos. | Neg. |
| β-catenin | Hom. Pos. | Hom. Pos. | Hom. Pos.** | Hom. Pos.** | Hom. Pos. |
| Vimentin | Neg. | Het. Pos. | Neg. | Hom. Pos. | Hom. Pos. |
| CD44 | Neg. | Neg. | Neg. | Neg. | Hom. Pos. |
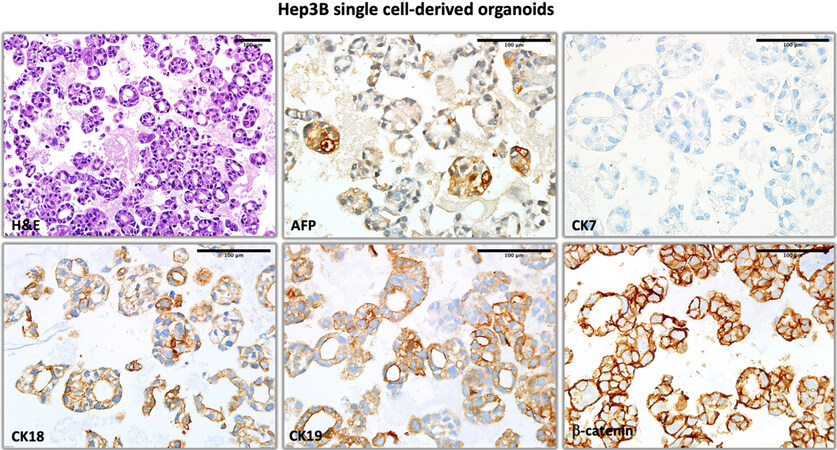
The heterogeneity of biomarker expression in HCC cell lines as observed under 3D conditions raised the question of cellular heterogeneity, although it is well known that many HCC cell lines display stem cell-like features with the ability to generate differentiated progeny from progenitor-like cells[16]. To test this hypothesis, we used single cell derived Hep3BTAp73β cells that we recently described[8]. 3D organoids obtained with single cell derived Hep3BTAp73β cells displayed cellular architectures and biomarker expression patterns that could not be distinguished from parental cells, as demonstrated with H&E staining, as well as AFP, CK7, CK18, CK19, and β-catenin immunostaining patterns [Figure 11].
DISCUSSION
Here we described experimental conditions to grow hepatoblast-like and mesenchymal-like HCC cells as 3D organoid-like structures. Our model that provided satisfactory results with five out of six cell lines tested is easy to perform and does not require specific reagent, except Matrigel. Successful 3D growth of three hepatoblast-like and two mesenchymal-like cell lines allowed us to compare their biomarker expression status, from which we are able to draw some conclusions on the common and unique characteristics of these two subgroups of cell lines.
All five 3D organoids that we were able to generate from HCC cell lines formed compacted structures similarly to organoids derived from primary HCC[6,7]. Hepatoblast-like organoids were more compacted than mesenchymal-like organoids reflecting their well-differentiated and poorly differentiated features, respectively.
Hepatoblast-like cell lines displayed homogenous expression of CK18 and β-catenin. They also commonly expressed E-cadherin, AFP, and A1AT, but heterogeneously. They were negative for CK7 expression, a marker for mature cholangiocytes[14]. Only Hep3B cells displayed 3D polarized cell structures and they expressed HepPar1 and glutamine synthetase, markers of mature hepatocytes. However, they also expressed CK19, a common marker for cholangiocytes and hepatobiliary progenitor cells[14]. Huh7 cells did not form polarized cell structures, but expressed hepatocyte markers arginase and A1AT, in addition to CK18. Huh7 cells, although homogeneously positive for AFP, did not express CK19. HepG2 cells, derived from a hepatoblastoma, heterogeneously positive for CK19 and AFP, but also E-cadherin and A1AT.
Colony-like structures formed by mesenchymal-like cell lines were loose and irregular. More interestingly, one of such cell lines, namely SNU475 was not able to form viable cell clusters in Matrigel. The remaining two mesenchymal-like cell lines shared homogenous expression of vimentin and β-catenin together with lack of expression of most hepatocyte markers as common features. However, both cell lines were able to form AFP-positive as well as E-cadherin-positive colonies. In addition, SNU449 displayed heterogeneous CK18 staining, but also homogenous staining for CD44.
Taken together, 3D culture of HCC cell lines belonging to two major subgroups revealed that such grouping of HCC cell lines is artificial since each cell line has its own cellular heterogeneity. In other terms, it is true that HCC cell lines, like primary tumors share many features in common. Nevertheless, they also have a private life with many unique features, as seen here with 3D organoid-like structures. Indeed, these cell lines express many markers heterogeneously as an indication of their ability to generate different progeny from the same ancestral cells. A large set of established HCC cell lines studied recently displayed mutation and transcriptomic profiles quite similar to those observed in primary tumors[5]. Therefore, the generation of progeny displaying different types of biomarker expression is not necessarily due to a genetic drift. However, we did not study here whether HCC cell lines reported here are genetically stable during cell culture. Such a hypothesis cannot be excluded. Another possibility for generation of heterogenous progeny under cell culture is the presence of stem/progenitor subpopulations in cell lines tested here. As reported previously, HCC cell lines harbor cancer stem cells able to generate differentiated progeny[16]. It will be interesting to further explore the mechanisms of cellular heterogeneity observed in this report.
Compared to primary tumor-derived organoids, cell line-derived organoids are much easier and less costly to obtain and maintain. The most noteworthy advantage of cell line-derived organoids is the ability to manipulate the expression status of selected genes either by ectopic expression or by gene knock-down or Cre-mediated gene inactivation that can be easily carried out under 2D monolayer culture conditions before testing them under 3D organoid-like conditions. Primary tumor derived organoids are usually maintained and expanded as 3D organoids which are less accessible to gene manipulation[6,7]. We have recently used Hep3B-derived clones to study TAp73 gene effects under 3D organoid conditions and demonstrated that this gene strongly affects biomarker expression and differentiation status of HCC cells[8]. Similar approaches could be applied to study the roles of selected genes in response of HCC cells to different therapeutic applications under tissue-like conditions. It is also possible to combine selected cancer and normal cell lines to generate heterotypic 3D organoid-like in vitro models to investigate tumor malignancy. Finally, their genomes are fully described in terms of structural alterations as well as expression profiles[5].
In conclusion, we describe here experimental conditions to obtain organoid-like structures from five different HCC cell lines representing hepatoblast-like and mesenchymal-like subgroups. These models will be useful as an alternative to monolayer cultures when studying proliferative, invasive, and metastatic abilities of HCC cells. Our detailed analysis of biomarker expression in five different organoid-like structures provides convincing evidence for highly specific phenotypic features of these cell lines, although they also share some common or subtype-restricted features.
DECLARATIONS
AcknowledgmentsWe thank the Izmir Biomedicine and Genome Center’s Optical Imaging Facility.
Authors’ contributionsStudy concept and design: Ozturk M, Oz O
Acquisition of data: Oz O, Iscan E, Batur T
Histological evaluation of samples: Oz O
Analysis and interpretation of data: Ozturk M, Oz O
Drafting of manuscript: Ozturk M, Oz O
Critical revision and final approval of the manuscript: Ozturk M
Obtained funding: Ozturk M
All authors have read and agreed to the published version of the manuscript.
Availability of data and materialsCell lines described here are available through major cell collections such as the American Type Culture Collection (ATCC). Detailed experimental conditions are available upon request.
Financial support and sponsorshipThis work was supported by TUBITAK grant no: 113S389, Turkish Academy of Sciences and in house funds from Izmir Biomedicine and Genome Center.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateHCC cell lines used in this study were obtained from ATCC. Ethics committee approval is not required.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
1. Cagatay T, Ozturk M. P53 mutation as a source of aberrant beta-catenin accumulation in cancer cells. Oncogene 2002;21:7971-80.
2. Fuchs BC, Fujii T, Dorfman JD, et al. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res 2008;68:2391-9.
3. Yuzugullu H, Benhaj K, Ozturk N, et al. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol Cancer 2009;8:90.
4. Hirschfield H, Bian CB, Higashi T, et al. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med 2018;50:e419.
5. Caruso S, Calatayud AL, Pilet J, et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology 2019;157:760-76.
6. Broutier L, Mastrogiovanni G, Verstegen MM, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 2017;23:1424-35.
7. Nuciforo S, Fofana I, Matter MS, et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep 2018;24:1363-76.
8. Iscan E, Ekin U, Yildiz G, et al. TAp73β Can Promote hepatocellular carcinoma dedifferentiation. Cancers (Basel) 2021;13:783.
9. Chua CW, Shibata M, Lei M, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol 2014;16:951-61, 1-4.
10. Lee SH, Hu W, Matulay JT, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018;173:515-28.e17.
11. Jing X, Li QK, Bedrossian U, Michael CW. Morphologic and immunocytochemical performances of effusion cell blocks prepared using 3 different methods. Am J Clin Pathol 2013;139:177-82.
12. Pan CC, Ho DM, Chen WY, Huang CW, Chiang H. Ki67 labelling index correlates with stage and histology but not significantly with prognosis in thymoma. Histopathology 1998;33:453-8.
13. Takai A, Fako V, Dang H, et al. Three-dimensional organotypic culture models of human hepatocellular carcinoma. Sci Rep 2016;6:21174.
14. Chougule P, Sumitran-holgersso S. Cytokeratins of the liver and intestine epithelial cells during development and disease. In: Hamilton G, editor. Cytokeratins - tools in oncology. InTech; 2012.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Oz O, Iscan E, Batur T, Ozturk M. 3D Organoid modelling of hepatoblast-like and mesenchymal-like hepatocellular carcinoma cell lines. Hepatoma Res 2021;7:60. http://dx.doi.org/10.20517/2394-5079.2021.43
AMA Style
Oz O, Iscan E, Batur T, Ozturk M. 3D Organoid modelling of hepatoblast-like and mesenchymal-like hepatocellular carcinoma cell lines. Hepatoma Research. 2021; 7: 60. http://dx.doi.org/10.20517/2394-5079.2021.43
Chicago/Turabian Style
Oz, Ozden, Evin Iscan, Tugce Batur, Mehmet Ozturk. 2021. "3D Organoid modelling of hepatoblast-like and mesenchymal-like hepatocellular carcinoma cell lines" Hepatoma Research. 7: 60. http://dx.doi.org/10.20517/2394-5079.2021.43
ACS Style
Oz, O.; Iscan E.; Batur T.; Ozturk M. 3D Organoid modelling of hepatoblast-like and mesenchymal-like hepatocellular carcinoma cell lines. Hepatoma. Res. 2021, 7, 60. http://dx.doi.org/10.20517/2394-5079.2021.43
About This Article
Copyright
Data & Comments
Data
 Cite This Article 20 clicks
Cite This Article 20 clicks













Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.