Genetics of alcohol-related hepatocellular carcinoma - its role in risk prediction
Abstract
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy, with increasing incidence worldwide. Alcohol-related cirrhosis (AC) accounts for 30% of the global incidence of HCC and HCC-related deaths. With the decline of hepatitis C virus (HCV) and decreasing HCV-related HCC, AC will soon become the leading cause of HCC. Excess alcohol consumption (> 80 g per day for > 10 years) increases the risk of HCC by 5-fold. However, only up to 35% of excessive drinkers develop cirrhosis and its associated HCC risk. Individual variation in susceptibility to HCC is known, but there is limited information to predict who among the patients is at high risk of progressing to HCC. Clinical risk factors for HCC include male gender, older age, severity of cirrhosis, obesity and presence of type 2 diabetes. In addition to ethnic variability in HCC risk, genetic variants are known to alter the risk of alcohol-related HCC. For example, single nucleotide polymorphisms in PNPLA3 (rs738409, C>G) and TM6SF2 (rs58542926, C>T) increase the risk of AC-related HCC, whereas HSD17B13 (T>A) reduces the risk for HCC. Studies have also confirmed PNPLA3 and TM6SF2 to be independent risk factors for AC-related (but not HCV-related) HCC. Combining genetic risk factors with phenotypic/clinical risk factors has been explored for stratification of patients for HCC development. Risk allele rs378409-G in PNPLA3 when combined with phenotypic/clinical risk factors (BMI, age, sex) has enabled HCC risk stratification of AC patients into low-, intermediate- and high-risk subgroups. Similarly, a combination of the two genetic variants PNPLA3-G and TM6SF2-T has been independently associated with risk of HCC onset. Using a polygenic risk score approach of incorporating several genetic variants, prognostic performance of polygenic risk score that included PNPLA3 rs378409 and TM6SF2 rs58542926 improved HCC prediction better than with either variant alone. Incorporating new variants and risk factors has the potential to build better algorithms/models to predict onset, early diagnosis and treatments for AC-related HCC. However, clinical usefulness of these approaches is yet to be determined.
Keywords
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy, with increasing incidence worldwide[1]. Despite screening programs in high-risk populations, long-term outcome is poor with a 5-year survival of 18%, representing the world’s third most lethal cancer. More specifically, the World Health Organization estimates that more than a million patients will die from liver cancer in 2030[2].
In almost 90% of cases, HCC occurs in the context of chronic liver disease, in particular, on the background of cirrhosis[1,3]. The underlying chronic liver disease promoting liver carcinogenesis varies geographically[1]. In Asia and sub-Saharan Africa, HCC is mostly caused by hepatitis B virus infection, while in the United States and Europe the current leading etiologies are hepatitis C virus (HCV) infection and alcohol-related cirrhosis (AC) followed by non-alcohol-related fatty liver disease (NAFLD)[4]. However, the advent of new direct-acting antiviral agents is expected to control HCV-related HCC in upcoming years, and AC will soon become the leading cause of HCC in most high-income countries[1]. Clinical risk factors for HCC occurrence include male gender, older age, severity of cirrhosis, obesity and presence of type 2 diabetes[5-7]. Clinical risk models have shown that the individual risk of HCC development is highly variable[6]. In addition, case-control and cancer database studies have highlighted the impact of ethnic background and a significant familial clustering[8,9]. For example, individuals of African and Hispanic ancestry are less likely to undergo curative therapies[10]. Overall, these observations strongly suggest that inherited genetic factors contribute to hepatocarcinogenesis.
Here, we review the current literature on risk factors, with a particular focus on genetic risk variants for alcohol-related HCC occurrence.
Epidemiology and characteristics of alcohol-related HCC
Alcohol-related HCC occurs infrequently in patients without pre-existing cirrhosis. Cirrhosis (of any etiology) is the single biggest risk factor for HCC development[3,11,12]. The annual incidence of HCC in patients with AC is nearly 3%[13]. The risk of developing AC and HCC parallels the amount of alcohol consumed daily and significantly increases above a threshold of 20 and 30 g for females and males, respectively[14,15]. Heavy alcohol drinking of more than 80 g per day for longer than 10 years increases the risk of HCC by 5-fold[16]. More specifically, AC accounts for 30% of the global incidence of HCC and HCC-related deaths, with marked geographical differences[17]. In Europe, HCC occurrence on the background of alcohol-related liver disease (ALD) varies from 20% in the south (e.g., Italy or Spain), to 63% in eastern countries. In the United States, alcohol accounts for 13% to 23% of HCC cases. Finally, the prevalence of alcohol-related HCC reaches 6% in the Middle East and 14% in North Africa[17,18]. The influence of alcohol consumption has also been highlighted by the impact of alcohol withdrawal on HCC development. Thus, a meta-analysis reported an annual reduction of HCC risk by 6%-7%[19].
However, only up to 10%-35% of excessive drinkers develop advanced fibrosis or cirrhosis and its associated HCC risk[20]. Interestingly, the role of alcohol consumption seems to be milder or even negligible compared to other environmental factors in the setting of HCC occurring in a non-fibrotic liver. For example, a recent case-control study observed that after adjustment of smoking habits and metabolic syndrome features, alcohol consumption was no longer independently associated with HCC in individuals with F0-F1 fibrosis stage[21].
HCC is often diagnosed at a later Barcelona Clinic Liver Cancer (BCLC) stage in patients with ALD and with a more severe underlying cirrhosis leading to a worse prognosis compared to other liver diseases[14,22]. A previous study reported that patients with alcohol-related HCC are often younger and are more frequently diagnosed with a multifocal or infiltrative/massive tumor compared to HCV-related HCC. However, this apparent greater cancer aggressiveness disappeared after adjusting for confounding factors, and prognosis was similar in ALD and HCV patients when stratified by BCLC stages[23]. Moreover, a recent study did not show significant differences in tumor characteristics between patients with AC- and NAFLD-related HCC[24]. Overall, the higher proportion of advanced BCLC stages observed in ALD/AC patients might only reflect a lower compliance with surveillance programs, rather than a greater tumor aggressiveness.
Limitations of screening strategies in patients with AC-related HCC
The American Association for the Study of Liver Diseases and European Association for the Study of the Liver recommend HCC surveillance in all cirrhotic patients using ultrasound, with or without alpha-fetoprotein determination, every 6 months[25,26]. This surveillance program has been shown to increase overall survival and improve the quality-adjusted life expectancy[27,28]. However, this periodic surveillance has been shown to be difficult to implement in daily clinical practice, ultimately leading to a significant prevalence of HCC detected at a more advanced stage[29]. Thus, more than 20% of HCC patients are also diagnosed with an unsuspected cirrhosis[30]. This phenomenon is even more pronounced in patients with ALD because AC is underdiagnosed due to their poor compliance in cancer surveillance programs[30]. Therefore, risk factors identified for AC are potential candidates for susceptibility to HCC.
Due to the aforementioned limitations, there is an urgent need for new detection strategies and the development of new highly sensitive, reliable, and easily accessible biomarkers that can either improve the early detection of HCC in high-risk patients with AC or identify individuals at risk of progressive ALD when liver fibrosis is incomplete and potentially reversible[31].
Individual variation in susceptibility to HCC is known, but there is limited information to predict who among the patients is at high risk of progressing to HCC. A better understanding of the contributing molecular, genetic and epigenetic factors is required to identify drivers of and therapeutic options for hepatocarcinogenesis.
Contribution of genetic variants to the prediction of alcohol-related HCC
The association of genetic variants with the risk of alcohol-related HCC has been reported. Earlier studies targeted genes with known functions, specifically genes known to operate in the pathogenesis of ALD and recently proposed to be part of a “5-hit working model” of disease progression leading to HCC[32]. These candidate genes, involved in hepatic alcohol metabolism [alcohol dehydrogenase (ADH), acetaldehyde dehydrogenase (ALDH), ethanol-induced cytochrome P450 (CYP2E1), CYP2E1-dependent microsomal ethanol oxidizing system (MEOS)] and affecting downstream mechanisms including oxidative stress [CYP2E1, glutathione S-transferase (GST), manganese superoxide dismutase (MnSOD), N-acetyltransferase 2, ectonucleotide pyrophosphatase/phosphodiesterase 1, homeostatic iron regulator (HFE)], endotoxin release [CD14, toll-like receptor 4 (TLR4)], immune function (TNFα, IFNg, IL-10, IL-1B, CD14) and fibrogenesis [TGFβ, angiotensin, leptin, metalloproteinases (MMPs), tissue inhibitors of MMPs (TIMPs)], have been extensively reviewed for their association to alter risk of ALD/AC and ALD-related HCC[12,33-35]. But results from most of these earlier studies could not be replicated or confirmed due to limitations in technology, small sample size, inappropriate study population and not accounting for underlying ethnic variability. The most widely known genetic mutations altering the risk of ALD and ALD-HCC are in the alcohol-metabolizing enzymes. These mutations alter the enzyme kinetics of alcohol dehydrogenase (ADH) and acetaldehyde dehydrogenase (ALDH)[35,36]. ADH1B rs1229984 induces ADH activity and acetaldehyde formation, whereas ALDH2*2 rs671 reduces ALDH activity, impairing its ability to clear acetaldehyde[35,37]. Carriage of both mutations results in the accumulation of toxic acetaldehyde levels with intense rise in arterial blood flow to the face, causing the well-known flushing and nausea. In East Asian populations with high prevalence of these mutations, this may result in reduced alcohol intake conferring protection against alcoholism[38]. Conversely, the risk of developing ALD and ALD-HCC increases in drinkers who carry one or both mutations[36].
Recent technological advances such as genome-wide association studies (GWAS) and next-generation sequencing have added to the growing field of genetic and epigenetic factors that modulate the risk for ALD/AC-related HCC. In recent years, a few single nucleotide polymorphisms (SNPs) have been discovered that are associated with risk of AC[39-41]. The single most commonly reproduced association with liver cirrhosis is the rs738409 SNP (p. I148M) in patatin-like phospholipase domain protein 3 (PNPLA3), which is also associated with increased HCC risk[42]. This C>G mutation is accompanied by a change from isoleucine to methionine at a conserved amino acid residue (I148M). Association of rs738409 (C>G) with increased risk of liver diseases has been confirmed in AC[40,41] and alcohol-related HCC[43,44]. Dose effect of the G-allele has been shown with ancestry-adjusted odds ratio (OR) increasing by 1.79 per G allele (P = 1.9 × 10-5) for ALD risk[45] and 1.77 (95%CI: 1.42-2.19, P = 2.78 × 10-7) per G allele for HCC[46]. The influence of this variant on HCC risk prediction revealed that the rs738409 (GG) genotype was an independent risk factor specifically for alcohol- (but not HCV-) related HCC[46]. Moreover, OR among the AC patients with HCC increased from 2.87 (95%CI: 1.61-5.10) in carriers of the CG genotype to 12.41 (95%CI: 6.99-22.03) in GG patients[43].
A study in a Chinese population showed that rs17401966 (A>G) in kinesin-like factor 1 B (KIF1B), a tumor suppressor gene, was associated with risk of HCC. The risk of HCC was higher in carriers of the AA genotype, compared to GG or AG, but only in the presence of alcohol (OR 2.36, 95%CI: 1.49-3.74), suggesting an additive gene-environment interaction between rs17401966 and alcohol consumption[47]. But this association has not been confirmed. Similarly, rs641738 (C>T) in membrane-bound O-acyltransferase 7 (MBOAT7), was identified as a risk locus for AC[40], but has yet to be replicated in other studies as a risk for AC/ALD or HCC.
Another SNP, rs58542926 (*/T) in transmembrane 6 superfamily 2 (TM6SF2), is strongly associated with the risk for HCC, particularly in patients with AC- and not HCV-related cirrhosis[44,48]. This variant was independently confirmed to be associated with HCC using a multivariate model adjusted for age, sex, BMI and diabetes (OR 2.5, 95%CI: 1.4-4.3)[48].
SNPs in hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13) are associated with decreased liver transaminases and liver injury[49]. In particular, recently identified splice variant SNP rs72613567 (T>A), resulting in loss of function and reduced enzyme activity, also showed (1) interactions with PNPLA3 rs738409 risk allele, with each rs72613567:TA allele lowering the increase in transaminase levels conferred by each PNPLA3 risk allele (I148M); and (2) allele dose-dependent association of rs72613567:TA with decrease in PNPLA3 mRNA expression[39]. Importantly, this rs72613566 (T>A) was associated with lower odds of alcohol- and non-alcohol-related liver diseases/cirrhosis as well as lower risk for HCC. The lower risk conferred by rs72613567 variant was PNPLA3 allele-dependent for AC and was confirmed in both men and women[50]. However, the rs72613567-associated lower risk for HCC was PNPLA3-dependent only in men[50].
It is suggested that for risk of AC/ALD-related HCC, PNPLA3 may be most relevant for the development of steatosis and ALD/AC, and TM6SF2 and MBOAT7 contributing towards HCC through inflammation-driven fibrosis[51]. Intriguingly, SNPs reported so far in PNPLA3, HSD17B13, TM6SF2 and MBOAT7 as being associated with AC/ALD are involved in lipid metabolism and processing, but their role in developing AC-ALD and HCC is yet to be clarified. Further investigations are required into the contribution of genetic factors individually and in combination with other variants, especially those influencing the effect on each other, such as PNPLA3 and HSD17B13. Understanding the roles of SNPs in the biology of liver disease is still in its infancy, and there is limited literature on specific functions of these recently identified SNPs. Although some SNPs are shared among different etiologies of cirrhosis and HCC, the role or interaction of these SNPs remains unclear in complex etiologies that co-exist with AC, such as viral hepatitis and NAFLD-related HCC. Further investigations are required to delineate the contribution of these SNPs to HCC development.
Risk stratification for AC/ALD-related HCC
Using a combination of genetic risk factors with other phenotypic/clinical risk factors has been explored for the identification and stratification of patients for HCC development[46,48,52-56]. Using the rs378409 risk variant in PNPLA3 in combination with other phenotype/clinical factors (BMI, age, sex) enabled HCC risk stratification of low-, intermediate- and high-risk AC patients (hazard ratio (HR 4.3, 95%CI: 2.7-6.4)[53]. Similarly, a combination of the two genetic variants PNPLA3-G and TM6SF2-T was independently associated with risk of HCC onset (HR 2.3, 95%CI: 1.5-3.4)[48]. Furthermore, the same study also reported that the number of HCC cases with carriage of both PNPLA3-G and TM6SF2-T risk alleles was significantly higher than carriers of only one risk allele in either SNP.
Most recently, a combination of all reported SNPs in AC, rs738409 (PNPLA3), rs6834314 (HSD17B13), rs58542926 (TM6SF2) and rs626283 (MBOAT7), when added to phenotypic factors (BMI, diabetes status, wine and coffee consumption), performed better [area under the curve (AUC) 0.748, 95%CI: 0.721-0.774], compared to only genetic factors (AUC 0.689, 95%CI: 0.660-0.717) or only phenotypic factors (AUC 0.681, 95%CI: 0.651-0.710), in stratifying drinkers with AC from drinkers with no liver disease[57]. It is encouraging that combining information on genetic variants with other risk factors can improve the identification of patients at risk. Furthermore, these genetic variants have also been shown to modulate severity of NAFLD (and its progression to steatohepatitis, fibrosis and cirrhosis) which commonly co-exists with alcoholic liver disease patients as “dual-etiology fatty liver disease” and accelerates liver injury[58,59].
Recently, the approach of incorporating several genetic variants in a so-called polygenic risk score (PRS) has been shown to be a successful strategy to improve the prediction of various complex phenotypes[60]. Thus, this method has been shown to outperform existing clinical models for the prediction of breast cancer with personalized recommendation on screening[61]. Of note, the addition of other risk factors into a global predictive model improves the overall performances compared to PRS alone[62]. At the transcriptional level, gene expression signatures gathering several dozens of genes (i.e., Prosigna and MammaPrint) have been included by the European Society for Medical Oncology to its clinical practice guidelines as prognostic and predictive tools to determine the benefit from chemotherapy[63].
The use of PRS to predict HCC occurrence in AC-related HCC patients is emerging[52,64]. More specifically, the prognostic performance of PRS including PNPLA3 rs738409 and TM6SF2 rs58542926 was higher than when considering PNPLA3 and TM6SF2 variants alone[52,64].
Several other SNPs have been identified as being associated with the risk of HCC, particularly with a viral etiology, but the literature is sparse regarding genetic variants specifically in relation to alcohol-related HCC. Similarly, contributions of molecular markers[65-67], somatic mutations[68], chromosomal instability and tumor microenvironment[69], and other regulatory components, such as mRNAs[70], noncoding RNAs[65,71], epigenetic[71,72] and mitoepigenetic factors[73] influencing the risk of alcohol-related HCC are few and overlap with other etiologies[69,71]. Last but not least, the role of the gut microbiota (fungi, bacteria and viruses), is another emerging factor influencing disease risk in ALD and ALD-HCC[74,75]. The gut microbiota also engages in alcohol metabolism, thereby altering the risk for ALD pathogenesis. Changes in the gut microbiome significantly correlates with alcohol consumption in human and experimental models, and there is evidence that alcohol and gut metabolites in ALD patients show carcinogenic effects[74], potentially increasing the risk of HCC.
Genomic studies have revealed several subclasses of HCC. Alcohol-related HCC is associated with CTNNB1 mutations (WNT-β-catenin signalling pathway); however, direct translation of molecular HCC subclasses into clinical management (i.e., personalized medicine) is yet to be achieved[76]. The recent success of checkpoint inhibitors in HCC has led to a renewed interest in immunological profiling of HCC and opportunities for personalized medicine. Recently, Sia et al.[77] analyzed the gene expression pattern of inflammatory cells in HCCs of almost 1000 patients. The authors identified a novel molecular class of tumors (in approximately 25% of patients) with an enriched inflammatory response characterized by overexpression of immune-related genes and high expression of PD1 and PD-L1 which may predict response to checkpoint inhibitor immunotherapy. A study by The Cancer Genome Atlas consortium performed multi-platform integrative molecular subtyping on 196 HCCs and found a similar subset of patients with high lymphocyte infiltration (in 22% of patients)[78]. Of note, the authors showed that the aforementioned CTNNB1 mutation was associated with a lack of immune infiltrate (so-called cold tumors), which has been observed by others[78,79]. In a recent first report of prospective genotyping of advanced HCC by next-generation sequencing, CTNNB1 mutations were associated with primary resistance to immune checkpoint inhibitors[80]. Patients exhibiting CTNNB1 mutations all had progressive disease as their best response and a shorter median survival compared to those without mutations (9.1 months vs. 15.2 months, respectively). Clearly, the immunological classification of alcohol-related HCCs will become increasingly important as immune-based therapies are added to the limited therapeutic options for patients with advanced disease.
With the discovery of new variants and risk factors, there is the potential to incorporate them for building prediction algorithms/models for AC/ALD-HCC onset, early diagnosis and treatments.
Clinical applications of risk-stratified AC/ALD-HCC patients
In terms of clinical application, patients identified by the above genetic modifiers to be at high risk of developing significant liver fibrosis may be prioritized for early referral to specialist care, with those at low risk remaining in primary care. These select high-risk patients can then be linked with resource-intensive multidisciplinary and evidence-based care involving hepatologists, psychiatrists, and addiction specialists to maximize their chance of obtaining abstinence. Indeed, when prolonged abstinence is achieved, it has been shown to lead to resolution of steatosis and inflammation and even fibrosis regression in some (but not all) patients[81,82]. Specialist care can also facilitate access to closer monitoring of liver fibrosis using non-invasive tests (e.g., transient elastography, magnetic resonance elastography) and prompt commencement of HCC surveillance (discussed below) when patients are diagnosed with cirrhosis.
Risk stratification for HCC surveillance
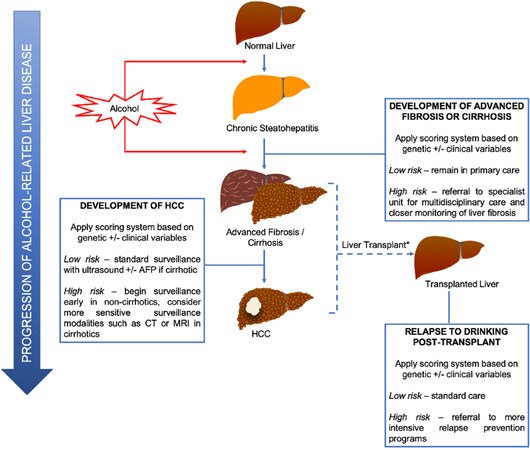
Aside from the prediction of patients at risk of advanced fibrosis or cirrhosis, genetic variants (e.g., PNPLA3, TM6SF2 and HSD17B13) can also help predict HCC development. As mentioned, these genes predisposing to alcohol-related HCC can be incorporated with other established risk factors for HCC (e.g., male sex, age and obesity) into a validated scoring system to risk-stratify patients for tailored HCC surveillance. Indeed, risk calculators for HCC development already exist for other liver diseases such as chronic hepatitis B and hepatitis C infection[83,84]. It is likely that the combination of several genetic variants (rather than any single SNP) with or without clinical variables into a score will be most predictive. Since the surveillance interval of 6 months for patients[85] was determined on the basis of estimated tumor doubling time (rather than tumor development risk), shortening intervals (e.g., to every 3 months) for ALD patients classified as high-risk may not result in improved outcomes[86]. However, risk calculators may help identify high-risk patients without cirrhosis, who should undergo surveillance (akin to surveillance of non-cirrhotic chronic hepatitis B patients) or those who should be surveyed with a more sensitive modality (e.g., computed tomography scan or magnetic resonance imaging). Conversely, risk scores may select out a low-risk group of patients with ALD who can safely forego surveillance, especially in resource-poor settings [Figure 1]. The development and application of risk scores using genetics needs to be explored further.
Figure 1. Potential clinical applications of genetic-based tests in alcoholic cirrhosis and related hepatocellular carcinoma. *It would be unethical to use genetic tests in the pre-transplant setting to determine if a patient should be offered liver transplantation. AFP: alpha-fetoprotein; CT: computed tomography; HCC: hepatocellular carcinoma; MRI: magnetic resonance imaging
Given the rise of alcohol-related HCC in the post-HCV era, it is imperative that more research be conducted in this area, providing a deeper understanding of the underlying risks and early diagnosis of HCC in patients with ALD. Possibilities exist of repurposing biomarkers and therapeutic agents for alcohol-related HCC identified/used for other etiologies.
Challenges of current risk prediction models for alcohol-related HCC
Risk prediction for alcohol-related HCC has been critically missing in the past due to lack of reproducible genetic studies. Recent discoveries on several genetic risk associations with AC have opened the field for using this information for risk prediction, not only for cirrhosis but also for alcohol-related HCC in patients with alcohol use problems.
The contribution of genetic variants, especially PNPLA3 rs738409, as potential predictors for AC-related HCC has been frequently discussed mainly because ORs often are > 2, calculated in retrospective cohorts[46]. However, modest to large ORs and extreme statistical significance do not automatically imply clinical relevance and other statistical metrics such as sensitivity, specificity and negative/positive predictive values might be more relevant[87]. Although variants in PNPLA3, TM6SF2 and more recently HSD17B13 modulate the risk of AC-related HCC, the use of these variants in HCC surveillance programs is currently not recommended[25,26].
Even though predictive models for HCC have generally been successful, they have limited clinical utility currently, especially the use of genetic-based factors identified in one ethnic population but used for prediction in another population. Although recent use of PRS for identification/stratification of at-risk patients is promising, one important limitation of PRS is their applicability in non-European ancestry populations. Indeed, most of the variants used in PRS have been identified in GWAS overwhelmingly conducted in individuals of European descent[88]. Therefore, the applicability of current PRS is not guaranteed[89]. Failure to include individuals from diverse ancestry will hamper the use of PRS in the multiethnic population seen in clinical practice[90]. At the level of gene expression, a 186-gene signature, initially developed to predict HCC recurrence in HCV-infected patients, has shown promising predictive ability for hepatocarcinogenesis in AC patients[91].
A particular challenge with alcohol-related liver diseases, including HCC, is the complication of alcohol dependence in these patients. Only select patients with alcohol-related HCC undergo liver transplantation (LT). After LT, up to 50% of patients relapse to drinking with 20% returning to harmful drinking with potential recurrence of liver disease[92]. The heritability of alcohol dependence has previously been estimated to be 25%-50%, and variants in genes encoding alcohol metabolism enzymes (ADH, ALDH) and GABA neurotransmission (GABRA2) have been shown to be associated with alcohol misuse[93]. Whether these same markers can predict (beyond current clinical markers) recidivism post-LT is currently unknown, so this presents an opportunity for further study. Transplanted patients identified to be at high risk of relapse can be preferentially referred for participation in multidisciplinary relapse prevention programs, which have been shown to be effective[94]. The prediction of relapsers post-LT will be increasingly important as transplant indications have recently expanded to include select patients with severe alcoholic hepatitis without significant prior abstinence[95].
Overall, PRS and gene expression signatures in combination with environmental risk factors have the potential to improve the prediction of alcohol-related HCC and pinpoint high-risk individuals. However, to date, evidence of clinical usefulness in this field is lacking. Thus, before genetic variation/expression can impact decision-making and be implemented in daily practice, it will need to be validated in large-scale prospective cohorts evaluating their clinical utility and cost-effectiveness[89,96]. Moreover, many physicians will require some training to interpret and communicate in a digestible manner the results of genetic testing and its current limitations[97].
Declarations
Authors’ contributionsLed the overall concept, design, structure, writing, submission and revision of the manuscript in consultation with all co-authors: Seth D
Risk stratification, HCC surveillance and treatment, Figure 1: Liu K
Introduction, epidemiology of AC-HCC, risk prediction: Trepo E, Verset G
Introduction, genetic variants, genetic variants risk prediction: Seth D
All authors critically reviewed the manuscript.
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2020.
REFERENCES
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
4. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604.
5. El-Serag HB, Tran T, Everhart JE. Diabetes increases the risk of chronic liver disease and hepatocellular carcinoma. Gastroenterology 2004;126:460-8.
6. N’Kontchou G, Paries J, Htar MTT, Ganne-Carrie N, Costentin L, et al. Risk factors for hepatocellular carcinoma in patients with alcoholic or viral C cirrhosis. Clin Gastroenterol Hepatol 2006;4:1062-8.
7. Zaman SN, Melia WM, Johnson RD, Portmann BC, Johnson PJ, et al. Risk factors in development of hepatocellular carcinoma in cirrhosis: prospective study of 613 patients. Lancet 1985;1:1357-60.
8. Yu MW, Chang HC, Liaw YF, Lin SM, Lee SD, et al. Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. J Natl Cancer Inst 2000;92:1159-64.
9. Hemminki K, Li X. Familial liver and gall bladder cancer: a nationwide epidemiological study from Sweden. Gut 2003;52:592-6.
10. Rich NE, Hester C, Odewole M, Murphy CC, Parikh ND, et al. Racial and ethnic differences in presentation and outcomes of hepatocellular carcinoma. Clin Gastroenterol Hepatol 2019;17:551-9.e1.
12. Stickel F, Schuppan D, Hahn EG, Seitz HK. Cocarcinogenic effects of alcohol in hepatocarcinogenesis. Gut 2002;51:132-9.
13. Ganne-Carrié N, Chaffaut C, Bourcier V, Archambeaud I, Perarnau JM, et al. Estimate of hepatocellular carcinoma incidence in patients with alcoholic cirrhosis. J Hepatol 2018;69:1274-83.
14. Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology 2004;127:S87-96.
15. Roerecke M, Vafaei A, Hasan OSM, Chrystoja BR, Cruz M, et al. Alcohol consumption and risk of liver cirrhosis: a systematic review and meta-analysis. Am J Gastroenterol 2019;114:1574-86.
17. Ganne-Carrié N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol 2019;70:284-93.
18. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol 2017;3:1683-91.
19. Heckley GA, Jarl J, Asamoah BO, G-Gerdtham U. How the risk of liver cancer changes after alcohol cessation: a review and meta-analysis of the current literature. BMC Cancer 2011;11:446.
20. Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011;141:1572-85.
21. Jean-Frédéric B, Adelaide D, Marie-Quitterie P, Aline M, Caroline B, et al. Diabetes, HBV infection and smoking are independent risk factors for developing hepatocellular carcinoma on non-fibrotic liver in the NoFLIC French multicenter case-control study. Hepatology 2015;62:276A.
22. Costentin CE, Mourad A, Lahmek P, Causse X, Pariente A, et al. Hepatocellular carcinoma is diagnosed at a later stage in alcoholic patients: results of a prospective, nationwide study. Cancer 2018;124:1964-72.
23. Bucci L, Garuti F, Camelli V, Lenzi B, Farinati F, et al; Italian Liver Cancer (ITA.LI.CA) Group; Italian Liver Cancer ITA LI CA Group. Comparison between alcohol- and hepatitis C virus-related hepatocellular carcinoma: clinical presentation, treatment and outcome. Aliment Pharmacol Ther 2016;43:385-99.
24. Kumar R, Goh BBG, Kam JW, Chang PE, Tan CK. Comparisons between non-alcoholic steatohepatitis and alcohol-related hepatocellular carcinoma. Clin Mol Hepatol 2020;26:196-208.
25. EASL. EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182-236.
26. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723-50.
27. Andersson KL, Salomon JA, Goldie SJ, Chung RT. Cost effectiveness of alternative surveillance strategies for hepatocellular carcinoma in patients with cirrhosis. Clin Gastroenterol Hepatol 2008;6:1418-24.
28. Bolondi L, Sofia S, Siringo S, Gaiani S, Casali A, et al. Surveillance programme of cirrhotic patients for early diagnosis and treatment of hepatocellular carcinoma: a cost effectiveness analysis. Gut 2001;48:251-9.
29. Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol 2020;72:250-61.
30. Singal AG, Yopp AC, Gupta S, Skinner CS, Halm EA, et al. Failure rates in the hepatocellular carcinoma surveillance process. Cancer Prev Res (Phila) 2012;5:1124-30.
31. Thursz M, Gual A, Lackner C, Mathurin P, Moreno C, et al. EASL Clinical Practice Guidelines: Management of alcohol-related liver disease. J Hepatol 2018;69:154-81.
32. Teschke R. Alcoholic liver disease: alcohol metabolism, cascade of molecular mechanisms, cellular targets, and clinical aspects. Biomedicines 2018;6:106.
33. Seitz HK, Stickel F. Risk factors and mechanisms of hepatocarcinogenesis with special emphasis on alcohol and oxidative stress. Biol Chem 2006;387:349-60.
35. Stickel F, Osterreicher CH. The role of genetic polymorphisms in alcoholic liver disease. Alcohol Alcohol 2006;41:209-24.
36. Homann N, Stickel F, Konig IR, Jacobs A, Junghanns K, et al. Alcohol dehydrogenase 1C*1 allele is a genetic marker for alcohol-associated cancer in heavy drinkers. Int J Cancer 2006;118:1998-2002.
37. Seitz HK, Stickel F. Molecular mechanisms of alcohol mediated carcinogenesis. Nat Rev 2007;7:599-612.
38. Dasgupta A. Genetic polymorphisms of alcohol metabolizing enzymes associated with protection from or increased risk of alcohol abuse. Alcohol, Drugs, Genes and the Clinical Laboratory 2017:107-16.
39. Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. Engl J Med 2018;378:1096-106.
40. Buch S, Stickel F, Trepo E, Way M, Herrmann A, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443-8.
41. Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet 2010;42:21-3.
42. Trepo E, Guyot E, Ganne-Carrie N, Degre D, Gustot T, et al. PNPLA3 (rs738409 C>G) is a common risk variant associated with hepatocellular carcinoma in alcoholic cirrhosis. Hepatology 2012;55:1307-8.
43. Salameh H, Raff E, Erwin A, Seth D, Nischalke HD, et al. PNPLA3 gene polymorphism is associated with predisposition to and severity of alcoholic liver disease. Am J Gastroenterol 2015;110:846-56.
44. Falleti E, Cussigh A, Cmet S, Fabris C, Toniutto P. PNPLA3 rs738409 and TM6SF2 rs58542926 variants increase the risk of hepatocellular carcinoma in alcoholic cirrhosis. Dig Liver Dis 2016;48:69-75.
45. Seth D, Daly AK, Haber PS, Day CP. Patatin-like phospholipase domain containing 3: a case in point linking genetic susceptibility for alcoholic and nonalcoholic liver disease. Hepatology 2010;51:1463-5.
46. Trépo E, Nahon P, Bontempi G, Valenti L, Falleti E, et al. Association between the PNPLA3 (rs738409 C\textgreaterG) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014;59.
47. Chen JH, Wang YY, Lv WB, Gan Y, Chang W, et al. Effects of interactions between environmental factors and KIF1B genetic variants on the risk of hepatocellular carcinoma in a Chinese cohort. World J Gastroenterol 2016;22:4183-90.
48. Nahon P, Cao Q, Guyot E, Rufat P, Sutton A, et al. O054 TM6SF2-T and PNPLA3-G genetic variants co-modulate the risk of hepatocellular carcinoma in caucasian patients with alcoholic cirrhosis. Inter-cohort validation in 1068 patients. J Hepatol 2015;62:S218-9.
49. Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131-8.
50. Stickel F, Lutz P, Buch S, Nischalke HD, Silva I, et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatology 2019. epub ahead of print. doi: 10.1002/hep.30996
51. Thursz M, Kamath PS, Mathurin P, Szabo G, Shah VH. Alcohol-related liver disease: Areas of consensus, unmet needs and opportunities for further study. J Hepatol 2019;70:521-30.
52. Yang J, Trépo E, Nahon P, Cao Q, Moreno C, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer 2019;144:533-44.
53. Guyot E, Sutton A, Rufat P, Laguillier C, Mansouri A, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol 2013;58:312-8.
55. Xu RH, Wei W, Krawczyk M, Wang W, Luo H, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155-61.
56. Yu JH, Suh YJ, Jin YJ, Yeo NY, Jang JW, et al. Prediction model for hepatocellular carcinoma risk in treatment-naive chronic hepatitis B patients receiving entecavir/tenofovir. Eur J Gastroenterol Hepatol 2019;31:865-72.
57. Whitfield JB, Schwantes-An TH, Cordell HJ, Darlay R, Masson S, et al. Can genotyping predict risk of alcohol-related cirrhosis in high-risk drinkers? Hepatology 2019;70:171A-2.
58. Anstee QM, Day CP. The genetics of nonalcoholic fatty liver disease: spotlight on PNPLA3 and TM6SF2. Semin Liver Dis 2015;35:270-90.
59. Anstee QM, Seth D, Day CP. Genetic factors that affect risk of alcoholic and nonalcoholic fatty liver disease. Gastroenterology 2016;150:1728-44.e7.
60. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018;19:581-90.
61. Maas P, Barrdahl M, Joshi AD, Auer PL, Gaudet MM, et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol 2016;2:1295-302.
62. Lee A, Mavaddat N, Wilcox AN, Cunningham AP, Carver T, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med 2019;21:1708-18.
63. Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015;26:v8-30.
64. Stickel F, Buch S, Nischalke HD, Weiss KH, Gotthardt D, et al. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am J Gastroenterol 2018;113:1475-83.
65. Scaggiante B, Kazemi M, Pozzato G, Dapas B, Farra R, et al. Novel hepatocellular carcinoma molecules with prognostic and therapeutic potentials. World J Gastroenterol 2014;20:1268-88.
66. Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, et al. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology 2011:483-90.
67. Sieghart W, Wang X, Schmid K, Pinter M, König F, et al. Osteopontin expression predicts overall survival after liver transplantation for hepatocellular carcinoma in patients beyond the Milan criteria. J Hepatol 2011;54:89-97.
68. Zhou SL, Zhou ZJ, Hu ZQ, Song CL, Luo YJ, et al. Genomic sequencing identifies WNK2 as a driver in hepatocellular carcinoma and a risk factor for early recurrence. J Hepatol 2019;71:1152-63.
69. Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: an update. World J Gastroenterol 2016;22:9069-95.
70. Ye QH, Qin LX, Forgues M, He P, Kim JW, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med 2003;9:416-23.
71. Wahid B, Ali A, Rafiqe S, Idrees M. New insights into the epigenetics of hepatocellular carcinoma. Biomed Res Int 2017;2017:1609575.
73. Lozano-Rosas MG, Chávez E, Aparicio-Cadena AR, Velasco-Loyden G, Sánchez VC. Mitoepigenetics and hepatocellular carcinoma. Hepatoma Res 2018;4:19.
74. Méndez-Sánchez N, Valencia-Rodríguez A, Vera-Barajas A, Abenavoli L, Scarpellini E, et al. The mechanism of dysbiosis in alcoholic liver disease leading to liver cancer. Hepatoma Res 2020;6:5.
75. Neuman MG, Seitz HK, French SW, Malnick S, Tsukamoto H, et al. Alcoholic-hepatitis, links to brain and microbiome: mechanisms, clinical and experimental research. Biomedicines 2020;8:63.
76. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018.
77. Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017;153:812-26.
78. Wheelar DA, Roberts LR. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327-41.e23.
79. Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouze E, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol 2017;67:727-38.
80. Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res 2019;25:2116-26.
83. Ganne-Carrie N, Layese R, Bourcier V, Cagnot C, Marcellin P, et al. Nomogram for individualized prediction of hepatocellular carcinoma occurrence in hepatitis C virus cirrhosis (ANRS CO12 CirVir). Hepatology 2016;64:1136-47.
84. Wong VW, Janssen HL. Can we use HCC risk scores to individualize surveillance in chronic hepatitis B infection? J Hepatol 2015;63:722-32.
85. Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358-80.
86. Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 2018;68:526-49.
87. Kraft P, Wacholder S, Cornelis MC, Hu FB, Hayes RB, et al. Beyond odds ratios--communicating disease risk based on genetic profiles. Nat Rev Genet 2009;10:264-9.
88. Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47:D1005-12.
89. Sugrue LP, Desikan RS. What are polygenic scores and why are they important? JAMA 2019;321:1820.
90. Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019;570:514-8.
91. Nakagawa S, Wei L, Song WM, Higashi T, Ghoshal S, et al. Molecular liver cancer prevention in cirrhosis by organ transcriptome analysis and lysophosphatidic acid pathway inhibition. Cancer Cell 2016;30:879-90.
92. DiMartini A, Day N, Dew MA, Javed L, Fitzgerald MG, et al. Alcohol consumption patterns and predictors of use following liver transplantation for alcoholic liver disease. Liver Transpl 2006;12:813-20.
93. Stickel F, Moreno C, Hampe J, Morgan MY. The genetics of alcohol dependence and alcohol-related liver disease. J Hepatol 2017;66:195-211.
94. Bjornsson E, Olsson J, Rydell A, Fredriksson K, Eriksson C, et al. Long-term follow-up of patients with alcoholic liver disease after liver transplantation in Sweden: impact of structured management on recidivism. Scand J Gastroenterol 2005;40:206-16.
95. Im GY, Cameron AM, Lucey MR. Liver transplantation for alcoholic hepatitis. J Hepatol 2019;70:328-34.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Liu K, Verset G, Trepo E, Seth D. Genetics of alcohol-related hepatocellular carcinoma - its role in risk prediction. Hepatoma Res 2020;6:42. http://dx.doi.org/10.20517/2394-5079.2020.25
AMA Style
Liu K, Verset G, Trepo E, Seth D. Genetics of alcohol-related hepatocellular carcinoma - its role in risk prediction. Hepatoma Research. 2020; 6: 42. http://dx.doi.org/10.20517/2394-5079.2020.25
Chicago/Turabian Style
Liu, Ken, Gontran Verset, Eric Trepo, Devanshi Seth. 2020. "Genetics of alcohol-related hepatocellular carcinoma - its role in risk prediction" Hepatoma Research. 6: 42. http://dx.doi.org/10.20517/2394-5079.2020.25
ACS Style
Liu, K.; Verset G.; Trepo E.; Seth D. Genetics of alcohol-related hepatocellular carcinoma - its role in risk prediction. Hepatoma. Res. 2020, 6, 42. http://dx.doi.org/10.20517/2394-5079.2020.25
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 3 clicks
Cite This Article 3 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.