Circulating tumor DNAs and non-coding RNAs as potential biomarkers for hepatocellular carcinoma diagnosis, prognosis and response to therapy
Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related deaths worldwide and despite improvement in therapeutic approaches, prognosis remains poor. This can be partly attributed to the fact that the majority of HCCs are diagnosed at intermediate or advanced stages. Availability of circulating biomarkers able to detect HCC at early stages could improve patients’ prognosis. At present, however, alpha fetoprotein or des-g-carboxyprothrombin are unable to reliably detect HCC at early stages and better circulating biomarkers are needed. Circulating tumor DNA (ctDNA) and non-coding RNAs (ncRNAs) are emerging as promising biomarkers to achieve the goal. Genetic and epigenetic alterations in ctDNA allow to pinpoint tumor-specific biomarkers, reveal tumor heterogeneity, help monitor tumor evolution over time and assess therapy efficacy. It remains to be fully evaluated the possibility of detecting these biomarkers at early tumor stages. Circulating ncRNAs are quantitative biomarkers with potential use in diagnostic, prognostic and predictive clinical settings. They may help to reveal HCC at early stages. However, because of heterogeneous and sometimes conflicting reported results, they still require validation and standardization of pre-analytical and analytical approaches before clinical applications could be envisaged.
Keywords
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and second leading cause of cancer-related deaths worldwide. Despite new therapeutic approaches, prognosis remains poor. According to the Barcelona Clinic Liver Cancer (BCLC) system[1], treatment options rely on cancer staging. Patients with an early stage HCC (BCLC stage A) can take advantage of curative treatments, such as tumor resection, ablation and liver transplantation. Although termed curative, risk of recurrence in post-surgical resection is about 70% at 5 years[2]. Patients who presents an intermediate (Stage B) or advanced (Stage C) HCC, about 70% of the total, can only benefit from palliative treatments, chemoembolization or sorafenib respectively, with survival in fewer than 10% of patients at 3 years[3-5].
Poor prognosis of HCC can be partly attributed to the fact that the majority of neoplasms are diagnosed at intermediate or advanced stages. Availability of blood biomarkers would be extremely important to improve early diagnosis in individual at risk or for a better management of prognosis and response to therapy in HCC patients. Among biomarkers presently in use, alpha fetoprotein (AFP) and des-γ-carboxy prothrombin (DCP) are the most commonly employed. AFP was the most widely used serum biomarker in HCC[6], but due to suboptimal sensitivity (55%-65%)[7-9], it is now employed to the monitoring of therapy effectiveness in HCC patients, together with ultrasound examination. DCP is a second biomarker utilized in HCC; although initially indicated as superior to AFP with a sensitivity of 92% and specificity of 93%[10], other studies showed suboptimal sensitivity (48%-62%)[11,12]. These studies signify that more effective biomarkers are needed for better management of HCC patients at various clinical phases.
Since 80%-90% of HCCs develop in a cirrhotic liver, a distinction between regenerative nodules and early HCC can be a challenge[13]. Albeit DCP exhibits the potential capability of differentiating HCC from non-malignant liver diseases[14], tissue biopsy remains the most dependable option for diagnostic purposes as well as for recognizing the molecular changes that characterize the tumor. However, tissue biopsy is invasive and associated with potential risks for the patients, it cannot be repeated and cannot be performed on patients with unresectable advanced HCC.
In recent years, liquid biopsy has become a valid alternative to overcome the above mentioned limitations. It is only modestly or not invasive at all and it offers the possibility of carrying out repeated tests over time. Moreover, it can be used for an early detection of tumors, for monitoring its growth dynamics, for evaluating the efficacy of treatments and for spotting tumor genetic heterogeneity and identifying mutations responsible for acquired resistance, becoming a highly promising approach for the clinical management of cancer patients. Liquid biopsy is the sampling and analysis of biological samples, such as blood, urine, saliva or stool, where nucleic acids originating from all or part of body districts can be found. In the presence of cancer, its derived materials, such as circulating tumor cells, cell-free tumor DNA (ctDNA) and microvesicles containing mRNAs, microRNAs (miRNAs) and proteins, are present in peripheral blood or other body fluids and can be measured through the use of specific tests [Figure 1]. This review is focused on ctDNA and circulating non coding RNAs, like miRNAs and long non-coding RNAs (lncRNAs), as potential biomarkers of HCC for early diagnosis, monitoring patients’ follow-up and assessing response to treatments.
Figure 1. Liquid biopsies and their clinical applications. A: Liquid biopsy is an approach for detecting and analyzing DNA and RNA in biological fluids, such as serum, plasma, urine and saliva. Being a minimally invasive procedure, it offers the possibility of performing repeated sampling over time, thus providing a practical method for patient surveillance. Blood plasma or serum from patients contain cancer derived material, such as CTCs, ctDNA, miRNAs and other RNAs; B: analysis of such DNA/RNA content can provide evidences on the presence of HCC at an early stage, assessment of prognosis and patients’ monitoring during and after therapy, thus helping the clinical management of patients at different phases of disease. CTC: circulating tumor cell; ctDNA: cell-free tumor DNA; ncRNA: non-coding RNA
ctDNA
The presence of free circulating DNA in serum/plasma has been used to reveal tumor-associated biomarkers, such as the increased abundance of cell-free DNA (cfDNA) in cancer patients or the presence of specific genetic or epigenetic alterations, which have been discovered in numerous types of cancer including HCC. Several studies have indeed proposed the cfDNA as a source of HCC biomarkers in diagnostic, prognostic or predictive clinical settings [Table 1].
Cell-free DNA as biomarker of hepatocellular carcinoma
| Target | Serum/plasma | Technology | Experimental setting | Clinical setting | Cohort comparison | Ref. |
|---|---|---|---|---|---|---|
| DNA abundance | Plasma | Fluorimeter | cfDNA levels | Diagnosis | HCC vs. HBV-related LF | [15] |
| DNA abundance | Serum | Branched DNA | cfDNA levels | Diagnosis | HCC vs. HL | [114] |
| DNA abundance | Plasma | Spectrophotometer | cfDNA levels | Prediction of radiotherapy response | HCC | [19] |
| DNA abundance | Plasma | Fluorimeter | cfDNA levels | Prognosis | HCC | [115] |
| GSTP1 | Serum | Real-time PCR | cfDNA levels | Diagnosis | HCC vs. HCV carriers | [16] |
| GSTP1 | Serum | Real-time PCR | cfDNA levels | Prognosis | HCC | [17] |
| hTERT | Plasma | Real-time PCR | cfDNA levels | Diagnosis | HCC vs. CHC + LC | [18] |
| hTERT | Plasma | Real-time PCR | cfDNA levels | Prognosis | HCC | [18] |
| APC, FHIT, p15, p16, E-cadherin | Plasma | MSP | Methylation | Diagnosis | HCC1 | [116] |
| APC, GSTP1, RASSF1A, SFRP1 | Plasma | MSRE-qPCR | Methylation | Diagnosis | HCC vs. HL | [21] |
| APC, GSTP1, RASSF1A, SFRP1 | Plasma | MSRE-qPCR | Methylation | Diagnosis | HCC vs. BLD | [21] |
| APC, GSTP1, RASSF1A, SFRP1 | Plasma | MSRE-qPCR | Methylation | Prognosis | HCC | [21] |
| Gene panel (12 genes) | Plasma | NGS | Methylation | Diagnosis | HCC vs. CLD + HL | [25] |
| p15, p16 | Plasma | MSP | Methylation | Diagnosis/prognosis | HCC vs. CLD + HL | [23] |
| SEPT9 | Plasma | MSP | Methylation | Diagnosis | HCC vs. LC | [117] |
| SOCS3 | Plasma | MSP | Methylation | Diagnosis/prognosis | HCC vs. LC, BLD + HL | [24] |
| UBE2Q1 | Serum | MSP | Methylation | Diagnosis | HCC vs. LC + CHB | [118] |
| Gene panel | Plasma | NGS | Mutations | Guiding therapy choice | HCC | [31] |
| Gene panel (574 genes) | Plasma | NGS | Mutations | Tumor heterogeneity and prognosis | HCC1 | [32] |
| Gene panel (58 genes) | Plasma/serum | NGS | Mutations | Patient monitoring | HCC1 | [27] |
| KRAS, NRAS | Plasma | BEAMing | Mutations | Guiding therapy choice | HCC | [31] |
| TERT | Plasma | ddPCR | Mutations | Diagnosis | HCC vs. LC | [119] |
| TERT | Plasma | ddPCR | Mutations | Prognosis | HCC | [119] |
| TERT, CTNNB1, TP53 | Plasma | NGS | Mutations | Prognosis | HCC1 | [120] |
| TP53 | Plasma | ddPCR | Mutations | Diagnosis | HCC vs. CLD + HL | [26] |
| TP53 | Plasma | COLD-PCR | Mutations | Patient monitoring | HCC | [121] |
| TP53, CTNNB1, TERT | Plasma | ddPCR | Mutations | Diagnosis | HCC1 | [28] |
| Genome-wide | Plasma | NGS | CNV | Diagnosis | HCC vs. CH + LC | [122] |
| Genome-wide | Plasma | NGS | SNV, CNV | Diagnosis | HCC vs. HL | [123] |
| Genome-wide | Plasma | NGS | SNV, CNV | Patient monitoring | HCC | [123] |
| Gene panel (54-70 genes) | Plasma | NGS | SNV, CNV, fusions | Guiding therapy choice | HCC | [29] |
cfDNA as biomarkers in plasma or serum in HCC
The level of plasma cfDNA was found significantly increased in patients affected by HCC, compared to individuals with liver fibrosis. A model that included this parameter together with patient age and AFP levels displayed 87.0% sensitivity and 100% specificity as diagnostic performance[15]. As a surrogate of abundance of circulating DNA, some studies evaluated the amount of cfDNA by quantifying specific circulating gene fragments. Iizuka et al.[16] found a significant increase in serum levels of the GSTP1 gene in HCC patients and found correlations with tumor grade and size. An increased GSTP1 gene in cfDNA was also associated with a shorter overall survival (OS) and metastasis occurrence[17]. Similarly, higher than normal levels of hTERT gene in plasma of HCC patients correlated with presence of advanced disease and shorter survival[18]. Quantification of cfDNA revealed its potential usefulness also for assessing therapy response. Reduction of plasma cfDNA after radiotherapy correlated with a better tumor response[19].
Quantification of circulating DNA is very easy to perform and inexpensive. However, this approach lacks specificity for type of cancer; in addition, levels of cfDNA can also increase in inflammatory conditions unrelated to cancer or in some physiological conditions, such as pregnancy. Furthermore, this analysis does not provide information about tumor genetic landscape and cannot reveal actionable targets. For these reasons, most investigations moved toward the detection of more specific genetic or epigenetic alterations in blood, as biomarkers for HCC.
Aberrant methylation of cfDNA in plasma or serum in HCC
Promoter methylation is a well-known mechanism for gene transcriptional repression. Aberrant methylation in promoters of cancer genes represents a tumor-specific event and its detection is potentially useful for the prediction or diagnosis of HCC. Concordance between aberrant methylations in tumor tissues and plasma is generally good, indicating that plasma could represent a tumor surrogate when tissue is not available. The field has been widely investigated and a meta-analysis of these studies has been published[20]. In the diagnostic setting, from the analysis of 150 plasma samples from patients with HCC, benign liver disease (including cirrhosis and chronic inactive hepatitis) and normal controls, Huang and co-workers found that the combined aberrant methylation of four genes (APC, GSTP1, RASSF1A and SFRP1) has a significant diagnostic value for HCC[21], confirming the results obtained in tumor tissues[22]. In particular, the combination analysis of plasma methylation levels of these genes allowed to discriminate HCC from both benign or normal controls, with a sensitivity of 84.7% (in both cases) and a specificity of 81.1% and 87.8% respectively[21].
For prognostic assessment, Wong et al.[23] evaluated the methylation status of p15 and p16 in tumor tissues, plasma, serum and buffy coat samples from HCC patients, non-HCC controls and healthy individuals and found promoter methylation in plasma or serum of 40%-50% HCC patients. Most of patients associated with gene methylation exhibited a poorer prognosis in comparison with patients negative for aberrant methylation[23]. The detection of methylation of APC and RASSF1A promoters was also associated with shorter OS in HCC patients and RASSF1A methylation was demonstrated to be an independent prognostic factor[21]. Very recently, another gene whose aberrant methylation detected in plasma was associated with patients’ poorer prognosis was SOCS3[24].
The analysis of panels of aberrantly methylated genes through the use of next generation sequencing (NGS) is expected to further improve sensitivity and specificity of aberrant methylation biomarkers. For example, targeted deep-sequencing of plasma DNA after bisulfite treatment could be used to simultaneously assess the methylation status of several targets. While Holmila et al.[25] identified two genes (VIM and FBLN1) whose promoters were differentially methylated in HCC using this approach, these areas of study have not been thoroughly investigated.
Compared to cfDNA abundance in plasma or serum, the detection of aberrant DNA methylations in cfDNA provides a more specific tumor biomarker, especially if a combination of multiple genes is employed. However, it still contains a limitation. Considering that aberrant methylation generally affects tumor suppressor genes, the analysis cannot reveal alterations in oncogenes potentially targets of specific therapies. With the development of more sophisticated approaches, the identification of tumor-specific genetic alterations has become feasible and has been applied to HCC.
Cancer gene mutations in plasma or serum cfDNA
An analysis of cancer gene mutations in serum or plasma of HCC patients was investigated in the diagnostic, prognostic and predictive settings.
In the diagnostic setting, R249S mutation of the TP53 gene is hallmark of aflatoxin B1 exposure, one of the major causes of HCC in certain geographic areas. Using droplet digital PCR the authors identified a higher prevalence of this mutation in plasma cfDNAs of HCC in Cameroonian and Central African patients in comparison with control subjects with and without liver disease (almost 25% of patients with HCC and 3%-9% of non-HCC subjects were R249S carriers), suggesting a potential use of this biomarker as an early risk factor for HCC in individuals exposed to aflatoxin B1[26]. Targeted deep sequencing was used to investigate several cancer genes involved in HCC. For example, the ultra-deep sequencing analysis of 58 cancer genes performed in 8 HCC tissues and paired plasma/serum samples revealed that 15 of the 21 somatic tumor mutations (71%) could also be detected in plasma/serum cfDNA[27], thus indicating the translational potential of this approach for HCC diagnosis. In another recent study, cancer alterations in four hot-spot regions of TP53, CTNNB1 and TERT genes were investigated in plasma cfDNA and corresponding tumor DNA from 48 HCC patients. Interestingly, the authors found that many gene alterations found in plasma DNA were different from those found in tumor tissues, an evidence of tumor heterogeneity[28]. Confirming tumor heterogeneity in HCC, a recent study analysed plasma cfDNA from 26 HCC patients for the presence of mutations in a large set of genes. Authors found tumor heterogeneity and evolution over time by tracking circulating mutation pattern in a patient who developed progression after capecitabine treatment[29].
The identification of gene mutations offers the possibility of identifying potential actionable alterations, useful for guiding treatment choice. It has been demonstrated that HCC harbouring mutant RAS exhibited a better clinical response to refametinib plus sorafenib, compared to wild-type RAS tumors[30]. Notably, in the course of the study aimed at detecting KRAS or NRAS mutations in plasma cfDNA of a large cohort of HCC patients, authors found other actionable mutations in EGFR, JAK2, BRAF, FLT3, PIK3CA, and cKIT, suggesting that available target therapies could potentially be effective in defined, albeit small, subsets of HCC patients[31]. These results highlight the usefulness of cfDNA analysis for identifying actionable targets and for stratifying patients according to the potentially most appropriate therapeutic approach.
The approach has also been employed to monitor therapy efficacy over time. The mutation analysis of 574 cancer genes applied to plasma cfDNA and matched HCC from four patients, demonstrated that 97% of tumor alterations were present in the blood and that it was possible to assess tumor progression, to track the possible sites of recurrence and understand tumor clonal dynamics in relation to sequential therapies. The possibility to track tumor dynamics from plasma analysis provides a valuable strategy for monitoring therapy efficacy and infer clinical outcomes[32], helping clinician modulate therapeutic approaches in a more rational and proper direction.
miRNAs
miRNAs are 20-24 nucleotides long RNAs. By interacting with homologous target mRNAs, they act by fine-tuning gene expression through a post-transcriptional mechanism. Each tissue exhibits a unique profile of miRNAs, which is altered in pathological conditions. In tumor tissues, miRNAs are aberrantly regulated and it has been demonstrated that some deregulated miRNAs can act as oncogenes and others as tumor suppressors[33]. The interest in circulating miRNAs as non-invasive tumor biomarkers surfaced when their presence was reported as stable molecules in serum or plasma of healthy individuals and cancer patients[34,35].
Approaches for detection and quantification of circulating miRNAs
The most common technologies employed to measure miRNA expression in biological samples include microarray, NGS, quantitative real-time PCR (RT-qPCR) and droplet digital PCR (ddPCR)[36-38]. Microarray and NGS technologies are suitable for screening and discovery purposes, qPCR and ddPCR remain the choices for validation and clinical tests development. Both microarrays and NGS provide high throughput analysis of miRNA expression profiles. Microarrays can quantify all the known miRNAs. NGS can also identify new miRNA species and differentiate closely related sequences. NGS can also detect miRNA length variation (isoforms of miRNA)[38]. qPCR and ddPCR are not high throughput technologies, but technology is relatively inexpensive, available in most laboratories and can offer higher sensitivity by exploiting amplification steps.
Among quantitative PCR approaches, ddPCR was shown to be superior to conventional real time qPCR for quantifying circulating miRNAs, as it allowed an easier absolute quantification of circulating RNAs without requiring an internal standard for normalization. Furthermore, ddPCR proved to be more tolerant than real time qPCR to the presence of inhibitors[39]. Finally, ddPCR generally exhibits a higher precision and reproducibility than real time qPCR, thus allowing an easier discrimination between cases and controls[37,40,41].
Circulating miRNAs for HCC diagnosis
Circulating miRNAs have been tested for their ability of discriminating HCC patients from control individuals [Table 2]. As shown in Table 2, however, it is evident that published studies are heterogeneous as they often differ for technical characteristics and experimental design. This heterogeneity makes it difficult to compare results and limits their transferability into applications of clinical interest.
Circulating miRNAs as diagnostic biomarkers of hepatocellular carcinoma
| miRNA | Expression | Body fluid | Experimental setting | Clinical setting | Cohorts comparison | Sample size | Sensitivity (%) | Specificity (%) | AUC | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-130b | Up | Serum | Single | Diagnosis | HCC vs. HL+ CHB | 57 vs. 30 + 29 | 87.7 | 81.4 | 0.91 | [124] |
| miR-29a-3p | Up | Serum | Single | Diagnosis | HCC vs. HL + LC (T) | 74 vs. 60 + 43 | N/A | N/A | 0.71 | [125] |
| Let-7f, miR-16, miR-21 | Down | Serum | Multiple | Diagnosis | HCC vs. HL | 90 vs. 60 | N/A | N/A | N/A | [47] |
| miR-101 | Up | Serum | Single | Diagnosis | HCC vs. HL | 25 vs. 20 | N/A | N/A | N/A | [126] |
| miR-101 | Down | Serum | Single | Diagnosis | HCC vs. HL | 67 vs. 30 | 76.1 | 70.0 | 0.79 | [53] |
| miR-101 | Down | Serum | Single | Diagnosis | HCC vs. LC | 67 vs. 61 | 95.5 | 90.2 | 0.98 | [53] |
| miR-101 | Down | Serum | Single | Diagnosis | HCC vs. HL + CH | 52 vs. 43 + 42 | 54.9 | 76.9 | 0.62 | [52] |
| miR-101 | Down | Serum | Single + AFP | Diagnosis | HCC vs. HL + CH | 52 vs. 43 + 42 | N/A | N/A | 0.85 | [52] |
| miR-101, miR-106b, miR-122, miR-195 | Down | Exosomes | Multiple | Diagnosis | HCC vs. CHB | 20 vs. 20 | N/A | N/A | N/A | [127] |
| miR-101-3p, miR-106b-3p | Serum | Multiple | Diagnosis | HCC vs. LC | 24 vs. 14 | 84.6 | 94.1 | 0.96 | [51] | |
| miR-101-3p, miR-1246, miR-106b-3p | Up | Plasma | Multiple | Diagnosis | HCC vs. HL | 22 vs. 11 | 100 | 100 | 1 | [51] |
| miR-101-3p, miR-1246, miR-106b-3p | Up | Plasma | Multiple | Diagnosis | HCC vs. HL + LC | 7 vs. 14 + 21 | N/A | N/A | N/A | [51] |
| miR-101-3p, miR-1246, miR-106b-3p | Up | Plasma | Multiple | Diagnosis | HCC vs. LC | 9 vs. 6 | 100 | 92.9 | 0.99 | [51] |
| miR-122 | Up | Serum | Single | Diagnosis | CH vs. HL | 30 vs. 10 | N/A | N/A | N/A | [56] |
| miR-122 | Up | Serum | Single | Diagnosis | HCC vs. HL | 30 vs. 10 | N/A | N/A | N/A | [56] |
| miR-122 | Down | Serum | Single | Diagnosis | LC vs. HL | 30 vs. 10 | N/A | N/A | N/A | [56] |
| miR-122 | Up | Single | Diagnosis | HCC vs. HL + CHB | 3423 vs. 1887 + 2403 | 68.0 | 73.3 | 0.77 | [128] | |
| miR-122 | Up | Serum | Single | Diagnosis | HCC vs. CHB | 71 vs. 45 | 77.6 | 57.8 | 0.63 | [45] |
| miR-122 | Up | Serum | Single | Diagnosis | HCC vs. HL | 71 vs. 34 | 81.6 | 83.3 | 0.87 | [45] |
| miR-122 | Down | Serum | Single | Diagnosis | CH vs. HL | 48 vs. 89 | 80.0 | 91.2 | 0.93 | [46] |
| miR-122 | Down | Serum | Single | Diagnosis | HCC vs. CHB | 101 vs. 89 | N/A | N/A | N/A | [46] |
| miR-122 | Up | Serum | Single | Diagnosis | HCC vs. HL | 101 vs. 89 | 70.7 | 69.1 | 0.79 | [46] |
| miR-122, let-7b | Up | Serum | Multiple | Diagnosis | HCC vs. HBV-related DN | 30 vs. 47 | 84.8 | 50.0 | 0.65 | [129] |
| miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-801 | Plasma | Multiple | Diagnosis | HCC vs. HL (V) | 196 vs. 66 | 83.2 | 93.9 | 0.94 | [49] | |
| miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-802 | Plasma | Multiple | Diagnosis | HCC vs. CHB (V) | 196 vs. 72 | 79.1 | 76.4 | 0.84 | [49] | |
| miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-803 | Plasma | Multiple | Diagnosis | HCC vs. LC (V) | 196 vs. 56 | 75.0 | 91.1 | 0.88 | [49] | |
| miR-122, miR-885-5p, miR-221, miR-181b | Serum | Multiple + AFP | Diagnosis | HCC vs. LC | 192 vs. 96 | N/A | N/A | 0.98 | [60] | |
| miR-122, miR-885-5p, miR-221, miR-181b | Serum | Multiple | Diagnosis | HCC vs. LC | 192 vs. 96 | N/A | N/A | 0.84 | [60] | |
| miR-122, miR-885-5p, miR-29b | Serum | Multiple + AFP | Diagnosis | HCC vs. HL | 192 vs. 96 | N/A | N/A | 1 | [60] | |
| miR-122, miR-885-5p, miR-29b | Serum | Multiple | Diagnosis | HCC vs. HL | 192 vs. 96 | N/A | N/A | 0.89 | [60] | |
| miR-122-5p | Up | Plasma | Single | Diagnosis | CHB vs. HL | 20 vs. 24 | N/A | N/A | N/A | [43] |
| miR-122-5p | Up | Plasma | Single | Diagnosis | HCC vs. HL | 20 vs. 28 | N/A | N/A | N/A | [43] |
| miR-122-5p | Up | Plasma | Single | Diagnosis | LC vs. HL | 20 vs. 22 | N/A | N/A | N/A | [43] |
| miR-122-5p | Up | Plasma | Single | Diagnosis | HCC vs. HL + LC | 7 vs. 14 + 21 | N/A | N/A | N/A | [51] |
| miR-122-5p | Up | Serum | Single | Diagnosis | HCC vs. LC | 24 vs. 14 | N/A | N/A | N/A | [51] |
| miR-122a | Down | Serum | Single | Diagnosis | HCC vs. HL | 85 vs. 85 | 70.6 | 67.1 | 0.71 | [130] |
| miR-122a | Down | Serum | Single + AFP | Diagnosis | HCC vs. HL | 85 vs. 85 | 87.1 | 98.8 | 0.94 | [130] |
| miR-1247-3p | Up | Serum | Single | Diagnosis | HCC vs. HL | 110 vs. 25 | N/A | N/A | N/A | [131] |
| miR-125a-5p | Down | Serum | Single | Diagnosis | HCC vs. HL + CHB | 120 vs. 164 + 91 | N/A | N/A | N/A | [76] |
| miR-125b | Down | Plasma | Single | Diagnosis | HCC vs. CHB | 64 vs. 63 | 93.8 | 85.7 | 0.96 | [58] |
| miR-125b | Down | Plasma | Single | Diagnosis | HCC vs. HL | 64 vs. 56 | 85.9 | 78.6 | 0.89 | [58] |
| miR-125b | Down | Plasma | Single | Diagnosis | HCC vs. LC | 64 vs. 59 | 89.1 | 88.1 | 0.96 | [58] |
| miR-125b, miR-223, miR-27a, and miR-26a | Down | Serum | Multiple + AFP | Diagnosis | HCC vs. HL | 90 vs. 60 | N/A | N/A | 0.87 | [50] |
| miR-125b, miR-27a | Down | Serum | Multiple + AFP | Diagnosis | HCC vs. HL | 90 vs. 60 | 80.0 | 87.2 | N/A | [50] |
| miR-125b-5p | Up | Plasma | Single | Diagnosis | CHB vs. HL | 20 vs. 24 | N/A | N/A | N/A | [43] |
| miR-125b-5p | Up | Plasma | Single | Diagnosis | HCC vs. HL | 20 vs. 28 | N/A | N/A | N/A | [43] |
| miR-125b-5p | Up | Plasma | Single | Diagnosis | LC vs. HL | 20 vs. 22 | N/A | N/A | N/A | [43] |
| miR-143 | Down | Serum | Single | Diagnosis | HCC vs. HL | 131 vs. 122 | 80.3 | 82.4 | 0.83 | [132] |
| miR-145 | Down | Serum | Single | Diagnosis | HCC vs. CHB | 85 vs. 50 | 88.2 | 78.0 | 0.85 | [133] |
| miR-148a | Down | Serum | Single | Diagnosis | HCC vs. HL + BLD | 76 vs. 55 + 62 | 67.7 | 59.2 | 0.67 | [71] |
| miR-148a, miR-148b, miR-152 | Down | Serum | Multiple | Diagnosis | HCC vs. BLD | 76 vs. 62 | 96.1 | 91.9 | 0.94 | [71] |
| miR-15b | Up | Serum | Single | Diagnosis | HCC vs. HL + CHB | 57 vs. 30 + 29 | 98.3 | 15.3 | 0.49 | [124] |
| miR-15b, miR-130b | Up | Serum | Multiple | Diagnosis | HCC vs. HL + CHB | 57 vs. 30 + 29 | 98.3 | 91.5 | 0.98 | [124] |
| miR-16 | Down | Serum | Single + AFP | Diagnosis | HCC vs. HL + CLD | 105 vs. 71 + 107 | 92.4 | 78.5 | N/A | [134] |
| miR-16, miR-195, miR-199a | Serum | Multiple | Diagnosis | HCC vs. HL + CLD | 105 vs. 71 + 107 | N/A | N/A | N/A | [134] | |
| miR-18, miR-221, miR-222, miR-224 | Up | Exosomes | Single | Diagnosis | HCC vs. LC | 20 vs. 20 | N/A | N/A | N/A | [127] |
| miR-18, miR-221, miR-222, miR-224 | Up | Exosomes | Single | Diagnosis | HCC vs. CHB + LC | 20 vs. 20 + 20 | N/A | N/A | N/A | [127] |
| miR-182 | Up | Serum | Single + AFP | Diagnosis | HCC vs. HL + BLD | 103 vs. 40 + 95 | 82.5 | 94.7 | [135] | |
| miR-182 | Up | Serum | Single | Diagnosis | HCC vs. HL + BLD | 103 vs. 40 + 95 | 78.6 | 91.6 | 0.91 | [135] |
| miR-182, miR-331-3p | Serum | Multiple + AFP | Diagnosis | HCC vs. HL + BLD | 103 vs. 40 + 95 | 93.2 | 95.8 | N/A | [135] | |
| miR-192-5p | Up | Serum | Single | Diagnosis | HCC vs. HL + LC (T) | 74 vs. 60 + 43 | N/A | N/A | 0.69 | [125] |
| miR-192-5p and miR-29a-3p | Up | Serum | Single | Diagnosis | HCC vs. HL (T) | 50 vs. 50 | N/A | N/A | N/A | [125] |
| miR-192-5p and miR-29a-3p | Up | Serum | Single | Diagnosis | HCC vs. HL (V) | 100 vs. 70 | N/A | N/A | N/A | [125] |
| miR-199a | Down | Serum | Single | Diagnosis | HCC vs. CH | 23 vs. 17 | 54.5 | 100 | 0.85 | [136] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. CH | 23 vs. 17 | 100 | 82.1 | 0.94 | [136] |
| miR-21 | Up | Serum | Single | Diagnosis | CHC vs. HL | 62 vs. 19 | 87.1 | 73.7 | 0.83 | [137] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. HL | 29 vs. 19 | N/A | N/A | N/A | [137] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. LC (V) | 175 vs. 78 | 80.8 | 72.9 | 0.81 | [59] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. CHB (V) | 175 vs. 64 | 76.9 | 85.7 | 0.79 | [59] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. HL (T) | 40 vs. 40 | [59] | |||
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. HL + CHB + LC (V) | 175 vs. 136 + 64 + 78 | 82.1 | 83.9 | 0.85 | [59] |
| miR-21 | Up | Single | Diagnosis | HCC vs. HL + CHB | 3423 vs. 1887 + 2403 | 86.6 | 79.5 | 0.88 | [128] | |
| miR-21 | Down | Serum | Single | Diagnosis | HCC vs. HL | 70 vs. 34 | N/A | N/A | N/A | [45] |
| miR-21 | Up | Plasma | Single + AFP | Diagnosis | HCC vs. HL + CH | 127 vs. 50 + 30 | 90.0 | 92.9 | 0.82 | [48] |
| miR-21 | Up | Plasma | Single | Diagnosis | HCC vs. HL + CH | 126 vs. 50 + 30 | 61.1 | 83.3 | 0.77 | [48] |
| miR-21 | Up | Exosomes | Single | Diagnosis | HCC vs. HL + CHB | 30 vs. 30 + 30 | N/A | N/A | N/A | [78] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. HL | 97 vs. 30 | N/A | N/A | N/A | [79] |
| miR-21 | Down | Serum | Single | Diagnosis | CH vs. HL | 48 vs. 89 | 80.0 | 95.6 | 0.91 | [46] |
| miR-21 | Down | Serum | Single | Diagnosis | HCC vs. CHB | 101 vs. 89 | [46] | |||
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. HL | 101 vs. 89 | 84.0 | 73.5 | 0.87 | [46] |
| miR-21 | Up | Serum | Single | Diagnosis | HCC vs. HL | 56 vs. 43 | 67.4 | 55.8 | 0.62 | [52] |
| miR-21, miR-122, miR-192 | Up | Serum | Multiple + AFP | Diagnosis | HCC vs. CHB | 118 vs. 100 | N/A | N/A | 0.94 | [61] |
| miR-21, miR-122, miR-192 | Up | Serum | Multiple | Diagnosis | HCC vs. CHB | 118 vs. 100 | N/A | N/A | 0.90 | [61] |
| miR-21, miR-122, miR-192 | Up | Serum | Multiple + AFP | Diagnosis | HCC vs. CHB + LC | 118 vs. 100 + 69 | N/A | N/A | 0.88 | [61] |
| miR-21, miR-122, miR-192 | Up | Serum | Multiple | Diagnosis | HCC vs. CHB + LC | 118 vs. 100 + 69 | N/A | N/A | 0.81 | [61] |
| miR-21, miR-122, miR-192 | Up | Serum | Multiple | Diagnosis | HCC vs. HL + CHB + LC | 118 vs. 119 + 100 + 69 | N/A | N/A | 0.85 | [61] |
| miR-21, miR-122, miR-192 | Up | Serum | Multiple + AFP | Diagnosis | HCC vs. LC | 118 vs. 69 | N/A | N/A | 0.88 | [61] |
| miR-21, miR-26a, miR-101 | Serum | Multiple + AFP | Diagnosis | HCC vs. HL + CH | 52 vs. 43 + 42 | 87.0 | 81.0 | 0.91 | [52] | |
| miR-218 | Down | Serum | Single + AFP | Diagnosis | HCC vs. HL + BLD | 156 vs. 64 + 98 | N/A | N/A | 0.91 | [73] |
| miR-218 | Down | Serum | Single | Diagnosis | HCC vs. HL + BLD | 156 vs. 64 + 98 | 66.7 | 69.1 | 0.73 | [73] |
| miR-22, miR-199a-3p | Serum | Multiple + AFP | Diagnosis | HCC vs. CHC | 192 vs. 96 | N/A | N/A | 0.98 | [60] | |
| miR-22, miR-199a-3p | Serum | Multiple | Diagnosis | HCC vs. CHC | 192 vs. 96 | N/A | N/A | 0.66 | [60] | |
| miR-221 | Up | Serum | Single | Diagnosis | CH vs. HL | 30 vs. 10 | N/A | N/A | N/A | [56] |
| miR-221 | Down | Serum | Single | Diagnosis | HCC vs. HL | 30 vs. 10 | N/A | N/A | N/A | [56] |
| miR-221 | Up | Serum | Single | Diagnosis | LC vs. HL | 30 vs. 10 | N/A | N/A | N/A | [56] |
| miR-221 | Up | Serum | Single | Diagnosis | HCC vs. HL | 46 vs. 50 | N/A | N/A | N/A | [72] |
| miR-221 | Up | Serum | Single | Diagnosis | HCC vs. HL | 45 vs. 45 | 93.3 | 77.8 | 0.94 | [138] |
| miR-221 | Up | Serum | Single + AFP | Diagnosis | HCC vs. HL | 45 vs. 45 | 96.5 | 88.0 | [138] | |
| miR-223 | Down | Serum | Single | Diagnosis | HCC vs. HL + CLD | 39 vs. 14 + 17 | 97.2 | 94.1 | 0.99 | [139] |
| miR-223 | Up | Serum | Single | Diagnosis | HCC vs. HL | 73 vs. 34 | N/A | N/A | N/A | [45] |
| miR-223 | Up | Serum | Single | Diagnosis | CH vs. HL | 48 vs. 89 | 80.0 | 75.0 | 0.88 | [46] |
| miR-223 | Up | Serum | Single | Diagnosis | HCC vs. HL | 101 vs. 89 | 80.0 | 76.5 | 0.86 | [46] |
| miR-223-3p | Down | Plasma | Single | Diagnosis | CHB vs. HL | 20 vs. 24 | N/A | N/A | N/A | [43] |
| miR-223-3p | Down | Plasma | Single | Diagnosis | HCC vs. HL | 20 vs. 28 | N/A | N/A | N/A | [43] |
| miR-223-3p | Down | Plasma | Single | Diagnosis | LC vs. HL | 20 vs. 22 | N/A | N/A | N/A | [43] |
| miR-223-3p | Down | Plasma | Single | Diagnosis | HCC vs. HL | 8 vs. 28 | N/A | N/A | N/A | [44] |
| miR-223-3p | Down | Plasma | Single | Diagnosis | LC vs. HL | 30 vs. 28 | N/A | N/A | N/A | [44] |
| miR-224 | Up | Plasma | Single | Diagnosis | HCC vs. HL | 20 vs. 20 | N/A | N/A | 0.91 | [140] |
| miR-224 | Up | Plasma | Single | Diagnosis | HCC vs. HL | 87 vs. 55 | 93.1 | 80.0 | 0.91 | [140] |
| miR-224 | Up | Plasma | Single | Diagnosis | HCC vs. HL | 33 vs. 22 | 87.7 | 86.3 | 0.91 | [140] |
| miR-224 | Up | Plasma | Single | Diagnosis | HCC vs. HL | 54 vs. 33 | 87.7 | 86.3 | 0.91 | [140] |
| miR-24-3p | Up | Serum | Single | Diagnosis | HCC vs. HL + CLD | 84 vs. 46 + 31 | N/A | N/A | 0.63 | [67] |
| miR-24-3p | Up | Serum | Single + AFP | Diagnosis | HCC vs. HL + CLD | 84 vs. 46 + 31 | N/A | N/A | 0.83 | [67] |
| miR-26a | Down | Serum | Single | Diagnosis | HCC vs. HL + CH | 52 vs. 43 + 42 | 75.0 | 70.0 | 0.76 | [52] |
| miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505 | Serum | Multiple | Diagnosis | HCC vs. CHB + LC (T) | 108 vs. 51 + 47 | 80.6 | 82.7 | 0.81 | [141] | |
| miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505 | Serum | Multiple | Diagnosis | HCC vs. CHB + LC (V) | 153 vs. 68 + 71 | 74.5 | 89.9 | 0.82 | [141] | |
| miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505 | Serum | Multiple | Diagnosis | HCC vs. HL + CHB + LC (T) | 108 vs. 51 + 51 + 47 | 80.6 | 85.5 | 0.82 | [141] | |
| miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505 | Serum | Multiple | Diagnosis | HCC vs. HL + CHB + LC (V) | 153 vs. 60 + 68 + 71 | 74.5 | 88.9 | 0.81 | [141] | |
| miR-30e | Down | Serum | Single | Diagnosis | HCC vs. HL + CLD | 39 vs. 14 + 17 | 91.7 | 70.5 | 0.93 | [139] |
| miR-331-3p | Up | Serum | Single | Diagnosis | HCC vs. HL + BLD | 103 vs. 40 + 95 | 79.6 | 92.6 | 0.89 | [135] |
| miR-331-3p | Up | Serum | Single + AFP | Diagnosis | HCC vs. HL + BLD | 103 vs. 40 + 95 | 91.2 | 92.6 | N/A | [135] |
| miR-335 | Down | Serum | Single | Diagnosis | HCC vs. HL + CH | 125 vs. 125 + 125 | N/A | N/A | N/A | [86] |
| miR-519d | Up | Exosomes | Single | Diagnosis | HCC vs. LC | 87 vs. 31 | N/A | N/A | 0.82 | [57] |
| miR-595 | Up | Exosomes | Single | Diagnosis | HCC vs. LC | 87 vs. 31 | N/A | N/A | 0.92 | [57] |
| miR-638 | Down | Exosomes | Single | Diagnosis | HCC vs. HL | 126 vs. 21 | N/A | N/A | N/A | [75] |
| miR-665 | Up | Exosomes | Single | Diagnosis | HCC vs. HL | 30 vs. 10 | N/A | N/A | N/A | [74] |
| miR-885-5p | Up | Serum | Single | Diagnosis | HCC + CHB + LC vs. HL | 46 + 23 + 26 vs. 24 | 90.5 | 79.2 | 0.94 | [142] |
| miR-939 | Up | Exosomes | Single | Diagnosis | HCC vs. LC | 87 vs. 31 | N/A | N/A | 0.84 | [57] |
| miR-96 | Up | Serum | Single | Diagnosis | HCC vs. CHB | 104 vs. 100 | 77.9 | 75.3 | 0.83 | [143] |
| miR-96 | Up | Serum | Single + AFP | Diagnosis | HCC vs. CHB | 104 vs. 100 | 83.6 | 82.4 | 0.88 | [143] |
| miR-96 | Up | Serum | Single | Diagnosis | HCC vs. CHB | 104 vs. 100 | 77.9 | 75.3 | 0.80 | [143] |
| miR-96 | Up | Serum | Single | Diagnosis | HCC vs. HL | 104 vs. 120 | N/A | N/A | N/A | [143] |
| miR-96 | Up | Serum | Single | Diagnosis | HCC vs. LC | 104 vs. 90 | N/A | N/A | N/A | [143] |
A first source of heterogeneity is given by the use of serum or plasma for measuring circulating miRNA levels. Albeit early studies reported that composition and levels of miRNAs in serum and plasma are similar[35], there are several examples that subsequently contradicted this idea. Heegaard et al.[42] tested miRNA levels in paired serum and plasma samples of lung cancer patients and they concluded that these apparently similar sources of circulating miRNAs exhibit very different miRNA levels. Supporting this conclusion in liver cancer patients, miR-223-3p was found consistently low in plasma of HCC patients[43,44] but the same miRNA was high in the serum[45,46]; miR-21 was found low in serum of HCC patients in two reports[45,47], but others reported miR-21 high in plasma of HCC patients[48,49]. Analysis of miR-125-b and miR-101 levels also displayed variances between serum and plasma: an upregulation of plasma miR-125-b levels was reported in HCC patients in comparison with healthy controls[43], while downregulation of the same miRNA was found in serum[50]; miR-101 levels were found high in plasma of HCC patients in comparison with healthy controls[51], whereas this miRNA was found downregulated in serum of HCC patients in comparison with healthy controls at least by two reports[52,53]. These several examples support the concept that differences between plasma and serum are common and should be taken in consideration when comparing results from different studies.
The use of plasma or serum is not the only source of variability of achieved results. An analysis of published studies strongly suggests that both pre-analytical and analytical procedures can affect results. Any change in tissue collection steps (like type of blood tubes, centrifugation strength and sample conservation) can generate differences in miRNA levels[54,55]. Considering the different hard-to-control sources of variability, it is not difficult to understand how uneven and sometimes even opposite results can easily derive.
In addition to technical reasons, aspects linked to experimental design can also be added to factors responsible for heterogeneity of results. The existence of various types of control populations represents indeed a source of variability and, in some cases, a limitation to the practical value of such results. In fact, for identifying useful biomarkers for the early detection of HCC it is very important to compare HCC not only with healthy controls but vs. cirrhotic patients, considering that 80%-90% of HCCs arise in this group of high-risk patients. A paradigmatic example is miR-122, a liver-specific miRNA whose level was found increased in serum/plasma of HCC patients in comparison with healthy patients[45,51], but studies found no significant differences when HCC patients were compared to cirrhotic or chronic hepatitis patients[51,56]. These findings indicate that increased circulating miR-122 levels likely reflect liver damage rather than the presence of an underlying HCC, indicating the importance of controls to draw conclusions. Among studies that produced results on differential circulating miRNAs between HCC vs. cirrhotic patients, Fornari et al.[57] found that serum miR-939, miR-595 and miR-494 could separate cirrhotic patients with and without HCC, performing better than AFP. Moshiri et al.[51] showed that the combination of three plasma miRNAs, miR-101-3p, miR-106b-3p and miR-1246, exhibited a high diagnostic accuracy in discriminating HCC from cirrhotic patients. Combination of two of the same miRNAs, miR-101-3p and miR-106b-3p, exhibited also an excellent diagnostic accuracy in serum of HCC vs. cirrhotic patients. Chen et al.[58] proved that plasma miR-125b could differentiate HCC from cirrhotic patients, and Guo et al.[59] found that serum miR-21 analysis had also a good diagnostic efficacy in discriminating HCC patients both from non-HCC populations or from cirrhotic patients. Analyses of miRNA panels have been used for discriminating cirrhotic patients with or without HCC. For example, two different miRNA panels have been employed to efficiently distinguish HCC patients from cirrhotic patients, one in serum (miR-122, miR-885-5p, miR-221, miR-181b[60]) and one in plasma (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-803[49]). More recently, Tat Trung et al.[61] showed that a miRNA panel including miR-21, miR-122 and miR-192 had a good diagnostic performance in discriminating HCC patients from other groups, in particular from cirrhotic and chronic hepatitis patients.
Another potential limitation of several published studies is the fact that they were based on the assumption that cell-free miRNA levels were altered as a consequence of their release by neoplastic cells, and tried to validate as circulating cancer biomarkers the same miRNAs deregulated in tumor tissues. This assumption may not be correct[62,63]. In fact, if we consider that ctDNA represents a small or very small fraction of cfDNA (approximately 0.1%-1% or less for non-metastatic tumors and 1%-10% for large metastatic tumors), it is difficult to conceive that cancer cells could instead release such a high amount of RNA to significantly change content and levels of specific circulating RNAs. While this consideration raises doubts about the source of circulating RNAs, a number of studies (see review by McAllister and Weinberg[64]) offer a possible explanation by indicating that cancer should be considered a systemic disease. In this view, tumor-associated systemic processes may unbalance the release of miRNAs from non-tumor cells and therefore change circulating RNA profiles. Thus, finding altered circulating miRNA profiles would represent evidence of a systemic pathophysiological process closely linked to the presence of a tumor, and such RNAs would then represent actual circulating tumor biomarkers, although largely not directly released by tumor cells. In this viewpoint, the numerous evidences that the altered levels of miRNAs in circulation do not necessarily reflect the miRNA deregulation found in cancer tissues would become plausible and results should be re-interpreted in accordance. In this context, an example is miR-101-3p, which was found downregulated in HCC tissues[65] but upregulated in plasma[51]. Other examples include miR-21, upregulated in HCC tissues[66] but in some cases they are reported to be downregulated in patients serum[45,47] or the previously mentioned miR-122, whose altered circulating level is predominantly a sign of hepatic injury[51,56].
The combination of biomarkers can potentially overcome the individual limitations. For this reason, miRNAs have been tested in association with AFP for improving test performance. Some studies indicated that the combination of miR-21 and AFP improved the discrimination between HCC patients vs. chronic hepatitis patients[48]. Meng et al.[67] showed that the combination of miR-24-3p and AFP allowed to better separate HCC from chronic liver disease affected patients and also a combination of miRNA panels with AFP provided a very good discriminating power between HCC vs. cirrhotic or chronic hepatitis patients[52,60,61].
Circulating miRNAs for HCC prognosis
Differences in median level of plasma/serum miRNAs as a cut-off value provided information about tumor stage and prognosis in HCC patients [Table 3]. Some studies have shown correlations of miRNA levels with pathological characteristics associated with prognosis. Members of the miR-148/152 family (miR-148a, miR-148b and miR-152) are important modulators of cell growth and progression of HCC[68-70]. Wang et al.[71] showed that low levels of miR-148a and miR-148b were significantly associated with tumor size and TNM stage, whereas low levels of miR-152 correlated with TNM stage. Additionally, the combination of circulating miR-148/152 family could discriminate HCC from non-malignant chronic liver diseases[71]. Other studies associated circulating miRNA levels with HCC patients’ prognoses. For example, Li et al.[72] showed that high serum levels of miR-221 correlated with tumor size, cirrhosis, tumor stage and with a lower OS in comparison with patient with low miR-221 expression levels, suggesting serum miR-221 as an independent risk factor for poor prognosis. A study by Yang et al.[73] reported that low serum levels of miR-218 were associated with clinic-pathological features such as tumor size, vascular invasion and TNM stage, as well as OS of patients. All these reports supported the potential role of circulating miRNAs in the assessment of prognosis in HCC. Studies described also the correlation between exosomal serum miRNA levels with pathological features and survival in HCC patients. For example, HCC patients with low levels of miR-665 showed a strong association with large tumour size (> 5 cm), local tumour invasion and metastases[74]; Shi et al.[75] showed that decreased levels of exosomal miR-638 had poor OS. Zheng et al.[76] reported that low levels of serum miR-125a-5p were associated with a lower OS compared with those exhibiting higher expression levels, and more recently Liu et al.[77] confirmed by a Kaplan-Meier analysis that HCC patients with lower serum miR-125b levels showed reduced time to recurrence and OS. Many studies involved the analysis of circulating miR-21. In one study, high levels of serum miR-21 were found correlated with cirrhosis and tumor stage[78], another revealed an association with metastasis[79]; Lee et al.[80] reported that exosomal miR-21 was an independent predictor of disease progression in HCC patients and high circulating levels of exosomal miRNA-21 were associated with lower OS and progression-free survival. These data support the role of miRNAs as potential prognostic biomarker in HCC.
Circulating miRNAs as prognostic biomarkers of hepatocellular carcinoma
| miRNA | Expression1 | Body fluid | Experimental setting | Clinical setting | Sample size | Kaplan-Meier analysis (P value) | Ref. |
|---|---|---|---|---|---|---|---|
| miR-101 | Down | Plasma | Single | Prognosis (DFS) | 163 | P < 0.001 | [144] |
| miR-122 | Down | Serum | Single | Prognosis (OS) | 122 | P < 0.01 | [145] |
| miR-1247-3p | Up | Serum | Single | Prognosis (OS) | 85 | P < 0.05 | [131] |
| miR-1247-3p | Up | Serum | Single | Prognosis (DFS) | 85 | P < 0.01 | [131] |
| miR-125a-5p | Down | Serum | Single | Prognosis (OS) | 120 | P < 0.01 | [76] |
| miR-125b | Down | Exosomes | Single | Prognosis (OS) | 128 | P < 0.01 | [77] |
| miR-143 | Down | Serum | Single | Prognosis (OS) | 131 | P < 0.05 | [132] |
| miR-148a | Down | Serum | Single | Prognosis (OS) | 76 | P < 0.001 | [71] |
| miR-152 | Down | Serum | Single | Prognosis (OS) | 76 | P < 0.05 | [71] |
| miR-192-5p | Up | Serum | Single | Prognosis (OS) | 74 | P < 0.01 | [125] |
| miR-192-5p | Up | Serum | Single | Prognosis (PFS) | 74 | P < 0.01 | [125] |
| miR-29a-3p | Up | Serum | Single | Prognosis (OS) | 74 | P < 0.01 | [125] |
| miR-29a-3p | Up | Serum | Single | Prognosis (PFS) | 74 | P < 0.05 | [125] |
| miR-21, lncRNA-ATB | Up | Exosomes | Single + lncRNA | Prognosis (OS) | 79 | P < 0.05 | [80] |
| miR-21, lncRNA-ATB | Up | Exosomes | Single + lncRNA | Prognosis (PFS) | 79 | P < 0.05 | [80] |
| miR-218 | Down | Serum | Single | Prognosis (OS) | 156 | P < 0.05 | [73] |
| miR-221 | Up | Serum | Single | Prognosis (OS) | 46 | P < 0.05 | [72] |
| miR-224 | Down | Serum | Single | Prognosis (OS) | 182 | P < 0.05 | [146] |
| miR-24-3p | Up | Serum | Single | Prognosis (OS) | 84 | P < 0.01 | [67] |
| miR-24-3p | Up | Serum | Single | Prognosis (DFS) | 84 | P < 0.01 | [67] |
| miR-638 | Down | Exosomes | Single | Prognosis (OS) | 126 | P < 0.01 | [75] |
| miR-96 | Up | Serum | Single | Prognosis (OS) | 49 | P < 0.05 | [143] |
Circulating miRNAs for prediction of HCC recurrence and treatment response
Many studies focused their attention on miRNAs ability to predict treatment response and monitor disease relapse after surgery or drug therapy [Table 4].
Circulating miRNAs as predictive biomarkers of therapy response in hepatocellular carcinoma
| miRNA | Expression in circulation1 | Body fluid | Experimental Setting | Clinical setting | Treatment | Sampling (pre/post therapy) | Ref. |
|---|---|---|---|---|---|---|---|
| miR-1246 | Up | Plasma | Single | Monitoring | LT | Pre/post | [82] |
| miR-182 | Up | Serum | Single | Monitoring | TACE | Pre/post | [135] |
| miR-331-3p | Up | Serum | Single | Monitoring | TACE | Pre/post | [135] |
| miR-122 | Up | Plasma | Single | Response prediction | TACE | Pre | [84] |
| miR-122 | Up | Plasma | Single | Response prediction | RFA | Pre | [147] |
| miR-181a-5p | Down | Serum | Single | Response prediction | Sorafenib | Pre | [89] |
| miR-200 | Up | Serum | Single | Response prediction | TACE | Pre | [87] |
| miR-21 | Up | Serum | Single | Response prediction | Resection | Pre | [79] |
| miR-21, miR-26a, miR-29a-3p | Plasma | Multiple | Response prediction | TACE | Pre | [148] | |
| miR-221 | Up | Serum | Single | Response prediction | Sorafenib | Pre | [88] |
| miR-26a | Down | Plasma | Single | Response prediction | Resection or RFA | Pre | [149] |
| miR-29a | Down | Plasma | Single | Response prediction | Resection or RFA | Pre | [149] |
| miR-339-5p | Down | Serum | Single | Response prediction | Sorafenib | Pre | [89] |
| miR-34a | Down | Serum | Single | Response prediction | Resection | Pre | [150] |
| miR-665 | Up | Serum exosomes | Single | Response prediction | Resection | Pre | [74] |
| miR-718 | Down | Serum exosomes | Single | Response prediction | LT | Pre | [151] |
| miR-1246 | Up | Plasma | Single | Responsive vs. non responsive | LT | Post | [82] |
| miR-148, miR-1246 | Up | Plasma | Multiple | Responsive vs. non responsive | LT | Post | [82] |
| miR-148a | Up | Plasma | Single | Responsive vs. non responsive | LT | Post | [82] |
| miR-148a, miR-148b, miR-152 | Down | Serum | Multiple | Responsive vs. non responsive | Resection | Post | [71] |
| miR-182 | Up | Serum | Single | Responsive vs. non responsive | TACE | Post | [135] |
| miR-221 | Down | Serum | Single | Responsive vs. non responsive | Sorafenib | Post | [88] |
| miR-331-3p | Up | Serum | Single | Responsive vs. non responsive | TACE | Post | [135] |
| miR-335 | Down | Serum | Single | Responsive vs. non responsive | TACE | Post | [86] |
| miR-423-5p | Down | Serum | Single | Responsive vs. non responsive | Sorafenib | Post | [152] |
| miR-122 | Down | Exosomal | Single | Responsive vs. non responsive | TACE | Pre/Post | [85] |
Surgery is the treatment of choice for early HCC, however relapse is common[81]. Levels of circulating miRNAs were studied in patients who underwent surgical resection, revealing a correlation with the post-operative survival. For example, subjects with low serum miR-21 levels had a 29% 5-year survival rate, whereas those with high expression had a 14.3% 5-year survival rate[79]. Wang et al.[71] showed that levels of serum miR-148/152 family decreased in case of relapse after surgery. Ng et al.[82] showed that miR-1246 was an independent predictor of OS and disease-free survival of HCC patients after liver transplant.
In patients at intermediate HCC stage, recommended first-line therapy is trans-arterial chemoembolization (TACE)[83], while sorafenib is the standard first line-systemic therapy for patients with advanced tumors (BCLC C)[81]. The association between circulating levels of miR-122 and treatment outcome after TACE was evaluated in two recent studies. Kim et al.[84] found that high plasma miR-122 expression levels could be predictive for early and overall TACE insufficient responses and refractoriness in HCC patients. Suehiro et al.[85] found that exosomal miR-122 expression levels were significantly decreased after TACE, especially in patients with cirrhosis, and suggest that the reduction in exosomal miR-122 levels may reflect a decrease in the liver function, rather than the anti-tumor effects of the procedure. Other miRNAs were studied in HCC patients treated with TACE. For example, lower serum miR-335 levels were associated with a shorter OS[86], while lower expression of miR-200 in HCC patients predicted a better prognosis in HCC patients treated with TACE[87]. Considering the data reported, circulating miRNAs could be associated with clinical outcome of HCC patients treated with TACE.
There are no biomarkers to predict response to sorafenib. A small number of studies evaluated whether circulating miRNAs could predict or anticipate therapy responsiveness. From the analysis of miR-221 levels in sera from HCC patients who received sorafenib, Fornari et al.[88] found that the treatment determined an increase of miR-221 only in responders. Moreover, analyzing miR-221 levels in sera from HCC patients before sorafenib treatment, lower miR-221-circulating levels were associated with better response to the drug[88]. Analysing serum miRNA profiles during sorafenib therapy, Nishida et al.[89] found that miR-181a-5p was decreased in non-responder patients compared to responders, suggesting this miRNA as a candidate serum biomarker for predicting response to sorafenib.
lncRNAs
lncRNAs are a class of > 200 nt RNA transcripts linked to the modulation of several pathways through various different molecular mechanisms[90,91]. Their expression differs between cancer and non-cancer tissues and their role in cancer biology is well recognized[92,93]. Expression deregulation of several lncRNAs has been reported in HCC[94] and abnormal levels of an increasing number of lncRNAs are being found in serum/plasma of HCC patients, suggesting their potential use as circulating tumor biomarkers. Table 5 summarizes the studies that have linked circulating lncRNAs to HCC. Technologies for detection and quantification of lncRNAs in biological samples are the same previously described for miRNAs.
Circulating long noncoding RNAs as biomarkers of hepatocellular carcinoma
| lncRNA | Expression | Body fluid | Experimental setting | Clinical setting | Cohorts comparison | Ref. |
|---|---|---|---|---|---|---|
| AF085935 | Up | Serum | Single | Diagnosis (early) | HCC vs. HL | [95] |
| AF085935 | Up | Serum | Single | Diagnosis (early) | HCC vs. HBV carriers | [95] |
| CTBP + LAMP2 + miR-16-2 + miR-21-5p | Serum | Multiple + miRNA | Diagnosis | HCC vs. HL + CHC | [153] | |
| CTBP + LAMP2 + miR-16-2 + miR-21-5p | Serum | Multiple + miRNA | Prognosis (PFS) | HCC | [153] | |
| DANCR | Up | Plasma | Single | Diagnosis | HCC vs. HL + CHB + LC | [105] |
| DANCR | Up | Plasma | Single | Prognosis | HCC | [105] |
| ENSG00000258332.1 + LINC00635 | Up | Serum | Multiple + AFP | Diagnosis | HCC vs. HL + LC + CHB | [96] |
| ENSG00000258332.1 + LINC00635 | Up | Serum | Single | Prognosis (OS) | HCC | [96] |
| HULC | Up | Plasma | Single | Diagnosis | HCC vs. HL | [104] |
| HULC | Up | Plasma | Single | Diagnosis | HCC vs. HL | [154] |
| HULC + LINC00152 | Up | Plasma | Multiple + AFP | Diagnosis | HCC vs. HL | [104] |
| JPX | Down | Plasma | Single + AFP | Diagnosis | HCC vs. HL | [101] |
| JPX | Down | Plasma | Single | Diagnosis | HCC vs. HL | [101] |
| JPX | Down | Plasma | Single | Prognosis | HCC | [101] |
| JUN + UCA1 | Up | Serum | Multiple | Diagnosis (early) | HCC vs. HL + CHC | [155] |
| JUN + UCA1 | Up | Serum | Multiple + AFP | Diagnosis (early) | HCC vs. HL + CHC | [155] |
| LRB1 | Up | Serum | Single + AFP/DCP | Diagnosis | HCC vs. HL | [102] |
| LRB1 | Up | Serum | Single | Prognosis (OS) | HCC | [102] |
| LINC00152 | Up | Plasma | Single | Diagnosis | HCC vs. HL | [104] |
| LINC00974 | Up | Plasma | Single | Diagnosis (early) | HCC vs. HL | [156] |
| LINC01225 | Up | Serum | Single | Diagnosis | HCC vs. HL | [157] |
| lnc-PCDH9-13:1 | Up | Saliva | Single | Diagnosis (early) | HCC vs. HL + HBV carrier + CHB + LC | [107] |
| LOC149086 + RP11-160H22.5 + XLOC_014172 | Up | Plasma | Multiple | Diagnosis | HCC vs. cancer free (T) | [158] |
| LOC149086 + RP11-160H22.5 + XLOC_014172 | Up | Plasma | Multiple | Diagnosis | HCC vs. cancer free (V) | [158] |
| PIVKAII + MALAT1 | Up | Plasma | Multiple + AFP | Diagnosis | HCC vs. HL | [159] |
| PVT1 + uc002mbe.2 | Up | Serum | Multiple | Diagnosis | HCC vs. HL | [100] |
| SNHG1 | Up | Plasma | Single | Diagnosis | HCC vs. HL + CHB + LC | [97] |
| SNHG1 | Up | Plasma | Single + AFP | Diagnosis | HCC vs. HL + CHB + LC | [97] |
| SPRY4-IT1 | Up | Plasma | Single + AFP | Diagnosis | HCC vs. HL | [160] |
| SPRY4-IT1 | Up | Plasma | Single | Diagnosis | HCC vs. HL | [160] |
| uc001ncr + AX800134 | Serum | Multiple | Diagnosis (early) | HCC vs. HL + HBV carriers | [161] | |
| uc003wbd | Up | Serum | Single | Diagnosis (early) | HCC vs. HL | [95] |
| uc003wbd | Up | Serum | Single | Diagnosis (early) | HCC vs. HBV carriers | [95] |
| UCA1 | Up | Serum | Single | Diagnosis | HCC vs. HL + CHC | [99] |
| UCA1 | Up | Serum | Single | Prognosis (PSF) | HCC | [103] |
| UCA1 + WRAP53 | Up | Serum | Multiple + AFP | Diagnosis | HCC vs. HL + CHC | [99] |
| UCA1 + WRAP53 | Up | Serum | Multiple | Diagnosis | HCC vs. HL + CHC | [99] |
| WRAP53 | Up | Serum | Single | Diagnosis | HCC vs. HL + CHC | [99] |
| WRAP53 | Up | Serum | Single | Prognosis (PSF) | HCC | [99] |
| XIST | Down | Plasma | Single | Prognosis | HCC | [101] |
| ZFAS1 | Up | Plasma | Single + AFP | Diagnosis | HCC vs. HL (T) | [98] |
| ZFAS1 | Up | Plasma | Single + AFP | Diagnosis | HCC vs. HL + CHB + LC (V) | [98] |
| ZFAS1 | Up | Plasma | Single | Diagnosis | HCC vs. HL (T) | [98] |
| ZFAS1 | Up | Plasma | Single | Diagnosis | HCC vs. HL + CHB + LC (V) | [98] |
A number of studies demonstrated that circulating lncRNAs could discriminate HCC from healthy controls or patients with non-malignant chronic liver diseases. In several cases, the studies involved detection of early HCCs, a crucial factor for the application of curative strategies. lncRNAs were evaluated either as single biomarkers or in combination. As single biomarkers, results indicated a sensitivity ranging from 51% to 92%. In this experimental setting, lncRNA - uc003wbd[95], ENSG00000258332.1[96], small nucleolar RNA host gene 1 (SNHG1)[97], zinc finger antisense 1 (ZFAS1)[98] were able to differentiate HCC patients from chronic hepatitis B virus (HBV) patients or healthy controls, urothelial carcinoma associated-1 (UCA1) or WRAP53[99] were significantly higher in HCC patients’ sera in comparison with chronic hepatitis C virus (HCV) patients or healthy volunteers. Diagnostic accuracy improved when lncRNAs were combined among them or with AFP and DCP. Combination of the lncRNAs plasmacytoma variant translocation 1 (PVT1) and uc002mbe.2 could discriminate early HCC patients from either HBV or HCV positive patients[100]. ZFAS1[98] or the expressed neighbor of XIST (Enox or JPX)[101] levels in combination with AFP could discriminate HCC from healthy individuals or chronic liver diseases patients with a better accuracy than each biomarker considered individually. The same was found for the combination of serum lncRNA uc007biz.1 (LRB1) with AFP and DCP biomarkers[102] or the combination of PVT1 and uc002mbe.2 with AFP[100].
Many circulating lncRNAs were also found to correlate with unfavourable pathologic features[96,97,100-105] and their association with prognosis was also evaluated. The increased levels of UCA1[106] or differentiation antagonizing non-protein coding RNA (DANCR)[105] or LRB1[102] were all associated to a poorer OS. Some of the studies investigated the combination of lncRNAs. The combined up-regulation of ENSG00000258332.1 and LINC00635[96], or of SNHG and UCA1[106] or low levels of JPX and X inactive-specific transcript (XIST)[101] significantly correlated with a poorer prognosis in HCC patients.
lncRNAs were also evaluated as biomarkers for monitoring tumor recurrence after surgery. In this clinical setting, it was found that patients with higher DANCR levels after surgery were prone to develop HCC recurrence[105]. It was also reported that the circulating levels of PCDH9-13:1 were significantly reduced after curative hepatectomy. However, it increased again in case of a relapse, suggesting that this lncRNA could be used to monitor patients after surgery[107]. Similarly, based on the decreased levels of SNHG1[97] or PVT1[100] after surgery, it was speculated that a subsequent increase of their expression might serve as biomarkers to monitor patients for HCC relapse.
Conclusion
Analysis of cell free DNA and RNA in body fluids, the so-called liquid biopsy, represents a very promising strategy for the early detection of cancer at an early stage or during monitoring of patients for the early detection of cancer relapse. The approach has the potential to significantly improve the clinical management of HCC cancer patients. Studies on circulating DNA/RNA in HCC originate from the need to identify more effective biomarkers than those currently in use, AFP and DCP. This review presents the results obtained so far in HCC. Although the specific studies still require further validation, overall they demonstrate a good sensitivity and specificity, higher than the current biomarkers and, once present limitations are over, can successfully find a valuable clinical use.
Although there are small amounts of nucleic acids in the circulation and only a fraction originate from tumor cells, the current technologies allow to pinpoint the changes induced by the presence of a tumor both qualitatively and quantitatively. Deep sequencing-based approaches for high-throughput and quantitative PCR analyses for targeted investigations demonstrated the appropriate ability to highlight such decisive traces of disease. However, it is clear that there are differences deriving from the analysis of DNA or RNA in the circulation.
From this point of view, DNA analysis is simpler and provides a straightforward interpretation: the presence, even in traces, of genetic or epigenetic alterations typically associated with a neoplastic disease provides a tangible sign of the presence of tumor cells. However, ctDNA analysis can also provide additional information. In fact, the technologies either based on NGS or ddPCR are quantitative and therefore allow to monitor the quantitative evolution of genetic or epigenetic alterations over time, thereby providing an assessment of therapy effectiveness. Furthermore, it is possible that distinct tumor clones may differentially respond to therapy. The mutational analysis of ctDNA offers the possibility of highlighting tumor heterogeneity, that tissue biopsy does not allow, thus making possible the detection of tumor clones differentially responsive to therapy. In this regard, identification of mutations in specific “actionable” genes offers the clinician the possibility of using targeted therapies if a molecular target is spotted. At present, the situation in HCC is limited, but auspiciously in evolution. Although sorafenib and regorafenib were the only few available options in advanced HCC until now, two new kinase inhibitors, namely lenvatinib and cabozantinib, have been recently approved by FDA for first-line and second-line treatment[108,109]. Unfortunately, an important limitation is that none of these drugs is associated with specific molecular alterations. As seen in other tumor types, it is however reasonable to expect that molecular subgroups of HCCs might be recognized for their differential responsiveness to specific targeted therapies. From these considerations, it is therefore clear that ctDNA analysis, which widely surpasses AFP and DCP circulating biomarkers in many aspects, can find clinical application for the management of HCC patients. It possibly presents a limitation, namely the possibility of detecting the presence of gene mutations in the early stages of disease, which can be difficult due to the very little amount of DNA released by small tumors. Overcoming this current limit will certainly be a goal of future research. Another limitation in HCC is the availability of a limited number of effective drugs against the most frequently mutated genes in HCC, such as catenin, as currently there are no target drugs capable of acting against the encoded oncoproteins.
The results from circulating RNA studies have probably a different conceptual meaning and so far the produced outcomes are not yet ready for clinical use. From a practical point of view, unlike genetic or epigenetic DNA-based tests that display characteristics intrinsically distinctive of neoplastic cells, in the case of circulating RNAs, they are also present in circulation of unaffected individuals and differences with cancer patients are exclusively quantitative. This indicates that for clinical application it will be necessary to define significance thresholds and appropriate methods to obtain reliable data. Quantitative PCR methods can be easily applied to diagnostic applications and, at present, ddPCR emerged as a robust method to quantify circulating miRNAs[37,39,40,110,111].
In summary, the analysis of circulating miRNA/lncRNA is potentially useful in different phases of the disease including early stages and the ddPCR method is optimally effective for performing the analysis. However, the approach is at present immature for clinical use both for methodological and scientific issues that need to be solved. Both pre-analytical and analytical procedures need to be standardized to guarantee solid results from independent laboratories[112]. On the scientific side, the levels of circulating RNAs show a normal variability among individuals. To diagnose HCC is essential to reaching an understanding of the variability in circulating miRNA/lncRNA levels not only in healthy individuals, but also and more importantly in patients affected by chronic liver diseases, such as chronic HBV/HCV infection, alcoholic and non-alcoholic fatty liver disease, steatohepatitis and cirrhosis. To this end, large study cohorts are needed[113]. Albeit often very exciting, results from the scientific literature are so far inconsistent. To change this status and open the way to translational applications, prospective well-designed large multicenter trials are needed.
Declarations
Authors’ contributionsConceived and designed the study: Callegari E, Sabbioni S, Negrini M
Wrote the manuscript: Guerriero P, Moshiri F, Lupini L, Callegari E, Negrini M
Read and approved the manuscript: All authors
Availability of data and materialsNot applicable.
Financial support and sponsorshipThis work was supported by funds from the Italian Association for Cancer Research and from the University of Ferrara to Negrini M.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2019.
REFERENCES
1. Llovet JM, Fuster J, Bruix J; Barcelona-Clinic Liver Cancer G. The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl 2004;10:S115-20.
2. Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg 2015;261:947-55.
3. Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-2.
4. Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival. Hepatology 2003;37:429-42.
5. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90.
6. Spangenberg HC, Thimme R, Blum HE. Serum markers of hepatocellular carcinoma. Semin Liver Dis 2006;26:385-90.
7. Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology 2009;137:110-8.
8. McMahon BJ, Bulkow L, Harpster A, Snowball M, Lanier A, et al. Screening for hepatocellular carcinoma in Alaska natives infected with chronic hepatitis B: a 16-year population-based study. Hepatology 2000;32:842-6.
9. Chen JG, Parkin DM, Chen QG, Lu JH, Shen QJ, et al. Screening for liver cancer: results of a randomised controlled trial in Qidong, China. J Med Screen 2003;10:204-9.
10. Volk ML, Hernandez JC, Su GL, Lok AS, Marrero JA. Risk factors for hepatocellular carcinoma may impair the performance of biomarkers: a comparison of AFP, DCP, and AFP-L3. Cancer Biomark 2007;3:79-87.
11. Grizzi F, Franceschini B, Hamrick C, Frezza EE, Cobos E, et al. Usefulness of cancer-testis antigens as biomarkers for the diagnosis and treatment of hepatocellular carcinoma. J Transl Med 2007;5:3.
12. Marrero JA, Su GL, Wei W, Emick D, Conjeevaram HS, et al. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology 2003;37:1114-21.
13. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018.
14. Lamerz R, Runge M, Stieber P, Meissner E. Use of serum PIVKA-II (DCP) determination for differentiation between benign and malignant liver diseases. Anticancer Res 1999;19:2489-93.
15. Yan L, Chen Y, Zhou J, Zhao H, Zhang H, et al. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis 2018;67:92-7.
16. Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res 2006;26:4713-9.
17. Tokuhisa Y, Iizuka N, Sakaida I, Moribe T, Fujita N, et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Cancer 2007;97:1399-403.
18. Piciocchi M, Cardin R, Vitale A, Vanin V, Giacomin A, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int 2013;7:1050-7.
19. Park S, Lee EJ, Rim CH, Seong J. Plasma cell-free DNA as a predictive marker after radiotherapy for hepatocellular carcinoma. Yonsei Med J 2018;59:470-9.
20. Zhang C, Li J, Huang T, Duan S, Dai D, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget 2016;7:81255-67.
21. Huang ZH, Hu Y, Hua D, Wu YY, Song MX, et al. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol 2011;91:702-7.
22. Hua D, Hu Y, Wu YY, Cheng ZH, Yu J, et al. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol 2011;91:455-60.
23. Wong IH, Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res 2000;6:3516-21.
24. Wei L, Huang Y, Zhao R, Zhang J, Liu Q, et al. Detection of promoter methylation status of suppressor of cytokine signaling 3 (SOCS3) in tissue and plasma from Chinese patients with different hepatic diseases. Clin Exp Med 2018;18:79-87.
25. Holmila R, Sklias A, Muller DC, Degli Esposti D, Guilloreau P, et al. Targeted deep sequencing of plasma circulating cell-free DNA reveals Vimentin and Fibulin 1 as potential epigenetic biomarkers for hepatocellular carcinoma. PLoS One 2017;12:e0174265.
26. Marchio A, Amougou Atsama M, Béré A, Komas NP, Noah Noah D, et al. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med 2018;18:421-31.
27. Labgaa I, Villacorta-Martin C, D’Avola D, Craig AJ, von Felden J, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 2018;37:3740-52.
28. Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer 2016;7:1907-14.
29. Ikeda S, Lim JS, Kurzrock R. Analysis of tissue and circulating tumor DNA by next-generation sequencing of hepatocellular carcinoma: implications for targeted therapeutics. Mol Cancer Ther 2018;17:1114-22.
30. Lim HY, Heo J, Choi HJ, Lin CY, Yoon JH, et al. A phase II study of the efficacy and safety of the combination therapy of the MEK inhibitor refametinib (BAY 86-9766) plus sorafenib for Asian patients with unresectable hepatocellular carcinoma. Clin Cancer Res 2014;20:5976-85.
31. Lim HY, Merle P, Weiss KH, Yau T, Ross P, et al. Phase II studies with refametinib or refametinib plus sorafenib in patients with RAS-mutated hepatocellular carcinoma. Clin Cancer Res 2018;24:4650-61.
32. Cai ZX, Chen G, Zeng YY, Dong XQ, Lin MJ, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer 2017;141:977-85.
33. Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci 2007;120:1833-40.
34. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008;141:672-5.
35. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8.
36. Thomson JM, Parker JS, Hammond SM. Microarray analysis of miRNA gene expression. Methods Enzymol 2007;427:107-22.
37. Miotto E, Saccenti E, Lupini L, Callegari E, Negrini M, et al. Quantification of circulating miRNAs by droplet digital PCR: comparison of EvaGreen- and TaqMan-based chemistries. Cancer Epidemiol Biomarkers Prev 2014;23:2638-42.
38. Wojcicka A, Swierniak M, Kornasiewicz O, Gierlikowski W, Maciag M, et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int J Biochem Cell Biol 2014;53:208-17.
39. Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem 2013;59:1670-2.
40. Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013;10:1003-5.
41. Mangolini A, Ferracin M, Zanzi MV, Saccenti E, Ebnaof SO, et al. Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark Res 2015;3:12.
42. Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012;130:1378-86.
43. Giray BG, Emekdas G, Tezcan S, Ulger M, Serin MS, et al. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep 2014;41:4513-9.
44. Oksuz Z, Serin MS, Kaplan E, Dogen A, Tezcan S, et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep 2015;42:713-20.
45. Qi P, Cheng SQ, Wang H, Li N, Chen YF, et al. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One 2011;6:e28486.
46. Xu J, Wu C, Che X, Wang L, Yu D, et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 2011;50:136-42.
47. Ge W, Yu DC, Li QG, Chen X, Zhang CY, et al. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin Lab 2014;60:427-34.
48. Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol 2012;56:167-75.
49. Zhou J, Yu L, Gao X, Hu J, Wang J, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 2011;29:4781-8.
50. Zuo D, Chen L, Liu X, Wang X, Xi Q, et al. Combination of miR-125b and miR-27a enhances sensitivity and specificity of AFP-based diagnosis of hepatocellular carcinoma. Tumour Biol 2016;37:6539-49.
51. Moshiri F, Salvi A, Gramantieri L, Sangiovanni A, Guerriero P, et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget 2018;9:15350-64.
52. Zhuang C, Jiang W, Huang D, Xu L, Yang Q, et al. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2016;40:386-96.
53. Xie Y, Yao Q, Butt AM, Guo J, Tian Z, et al. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther 2014;15:1248-55.
54. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492-7.
55. Zheng XH, Cui C, Zhou XX, Zeng YX, Jia WH. Centrifugation: an important pre-analytic procedure that influences plasma microRNA quantification during blood processing. Chin J Cancer 2013;32:667-72.
56. El-Garem H, Ammer A, Shehab H, Shaker O, Anwer M. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol 2014;6:818-24.
57. Fornari F, Ferracin M, Trerè D, Milazzo M, Marinelli S, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One 2015;10:e0141448.
58. Chen S, Chen H, Gao S, Qiu S, Zhou H, et al. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol Res 2017;47:312-20.
59. Guo X, Lv X, Lv X, Ma Y, Chen L, et al. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017;8:44050-8.
60. Zekri AN, Youssef AS, El-Desouky ED, Ahmed OS, Lotfy MM, et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol 2016;37:12273-86.
61. Tat Trung N, Duong DC, Tong HV, Hien TTT, Hoan PQ, et al. Optimisation of quantitative miRNA panels to consolidate the diagnostic surveillance of HBV-related hepatocellular carcinoma. PLoS One 2018;13:e0196081.
62. Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol 2014;8:819-29.
63. Cookson VJ, Bentley MA, Hogan BV, Horgan K, Hayward BE, et al. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell Oncol (Dordr) 2012;35:301-8.
64. McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16:717-27.
65. Su H, Yang JR, Xu T, Huang J, Xu L, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 2009;69:1135-42.
66. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61.
67. Meng FL, Wang W, Jia WD. Diagnostic and prognostic significance of serum miR-24-3p in HBV-related hepatocellular carcinoma. Med Oncol 2014;31:177.
68. Zhang JP, Zeng C, Xu L, Gong J, Fang JH, et al. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene 2014;33:4069-76.
69. Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 2010;52:60-70.
70. Zhang JG, Shi Y, Hong DF, Song M, Huang D, et al. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/beta-catenin pathway. Sci Rep 2015;5:8087.
71. Wang F, Ying H, He B, Pan Y, Sun H, et al. Circulating miR-148/152 family as potential biomarkers in hepatocellular carcinoma. Tumour Biol 2016;37:4945-53.
72. Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun 2011;406:70-3.
73. Yang L, Xu Q, Xie H, Gu G, Jiang J. Expression of serum miR-218 in hepatocellular carcinoma and its prognostic significance. Clin Transl Oncol 2016;18:841-7.
74. Qu Z, Wu J, Wu J, Ji A, Qiang G, et al. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017;8:80666-78.
75. Shi M, Jiang Y, Yang L, Yan S, Wang YG, et al. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem 2018;119:4711-6.
76. Zheng J, Zhou Z, Xu Z, Li G, Dong P, et al. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol Med Rep 2015;12:1584-90.
77. Liu W, Hu J, Zhou K, Chen F, Wang Z, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther 2017;10:3843-51.
78. Wang H, Hou L, Li A, Duan Y, Gao H, et al. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int 2014;2014:864894.
79. Wang X, Zhang J, Zhou L, Lu P, Zheng ZG, et al. Significance of serum microRNA-21 in diagnosis of hepatocellular carcinoma (HCC): clinical analyses of patients and an HCC rat model. Int J Clin Exp Pathol 2015;8:1466-78.
80. Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, et al. Circulating exosomal non-coding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer 2018; doi: 10.1002/ijc.31931.
81. Liver EAftSot. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol 2018;69:182-236.
82. Ng KT, Lo CM, Wong N, Li CX, Qi X, et al. Early-phase circulating miRNAs predict tumor recurrence and survival of hepatocellular carcinoma patients after liver transplantation. Oncotarget 2016;7:19824-39.
83. European Association for Study of L, European Organisation for R, Treatment of C. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Eur J Cancer 2012;48:599-641.
84. Kim SS, Nam JS, Cho HJ, Won JH, Kim JW, et al. Plasma micoRNA-122 as a predictive marker for treatment response following transarterial chemoembolization in patients with hepatocellular carcinoma. J Gastroenterol Hepatol 2017;32:199-207.
85. Suehiro T, Miyaaki H, Kanda Y, Shibata H, Honda T, et al. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett 2018;16:3267-73.
86. Cui L, Hu Y, Bai B, Zhang S. Serum miR-335 Level is Associated with the Treatment Response to Trans-Arterial Chemoembolization and Prognosis in Patients with Hepatocellular Carcinoma. Cell Physiol Biochem 2015;37:276-83.
87. Liu M, Liu J, Wang L, Wu H, Zhou C, et al. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One 2014;9:e109347.
88. Fornari F, Pollutri D, Patrizi C, La Bella T, Marinelli S, et al. In hepatocellular carcinoma miR-221 modulates Sorafenib resistance through inhibition of caspase-3 mediated apoptosis. Clin Cancer Res 2017; doi: 10.1158/1078-0432.CCR-16-1464.
89. Nishida N, Arizumi T, Hagiwara S, Ida H, Sakurai T, et al. MicroRNAs for the Prediction of Early Response to Sorafenib Treatment in Human Hepatocellular Carcinoma. Liver Cancer 2017;6:113-25.
90. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013;14:699-712.
91. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 2018;19:143-57.
93. Tang Y, Cheung BB, Atmadibrata B, Marshall GM, Dinger ME, et al. The regulatory role of long noncoding RNAs in cancer. Cancer Lett 2017;391:12-9.
94. DiStefano JK. Long noncoding RNAs in the initiation, progression, and metastasis of hepatocellular carcinoma. Noncoding RNA Res 2017;2:129-36.
95. Lu J, Xie F, Geng L, Shen W, Sui C, et al. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumour Biol 2015;36:3231-6.
96. Xu H, Chen Y, Dong X, Wang X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev 2018;27:710-6.
97. Gao S, Xu X, Wang Y, Zhang W, Wang X. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol Med Rep 2018;18:3305-13.
98. Luo P, Liang C, Zhang X, Liu X, Wang Y, et al. Identification of long non-coding RNA ZFAS1 as a novel biomarker for diagnosis of HCC. Biosci Rep 2018;38.
99. Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, et al. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res 2016;168:134-45.
100. Yu J, Han J, Zhang J, Li G, Liu H, et al. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltimore) 2016;95:e4436.
101. Ma W, Wang H, Jing W, Zhou F, Chang L, et al. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2017;41:163-70.
102. Wang ZF, Hu R, Pang JM, Zhang GZ, Yan W, et al. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol Lett 2018;16:1593-601.
103. Zheng ZK, Pang C, Yang Y, Duan Q, Zhang J, et al. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J Int Med Res 2018;46:348-56.
104. Li J, Wang X, Tang J, Jiang R, Zhang W, et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem 2015;37:687-96.
105. Ma X, Wang X, Yang C, Wang Z, Han B, et al. DANCR Acts as a Diagnostic Biomarker and Promotes Tumor Growth and Metastasis in Hepatocellular Carcinoma. Anticancer Res 2016;36:6389-98.
106. Li J, Gao J, Kan A, Hao T, Huang L. SNHG and UCA1 as prognostic molecular biomarkers in hepatocellular carcinoma: recent research and meta-analysis. Minerva Med 2017;108:568-74.
107. Xie Z, Zhou F, Yang Y, Li L, Lei Y, et al. Lnc-PCDH9-13:1 Is a Hypersensitive and Specific Biomarker for Early Hepatocellular Carcinoma. EBioMedicine 2018;33:57-67.
108. Kudo M, Finn RS, Qin S, Han KH, Ikeda K, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163-73.
109. Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, et al. Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N Engl J Med 2018;379:54-63.
110. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10.
111. Ferracin M, Lupini L, Salamon I, Saccenti E, Zanzi MV, et al. Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget 2015;6:14545-55.
112. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem 2015;61:56-63.
113. Lu LC, Hsu CH, Hsu C, Cheng AL. Tumor Heterogeneity in Hepatocellular Carcinoma: Facing the Challenges. Liver Cancer 2016;5:128-38.
114. Chen K, Zhang H, Zhang LN, Ju SQ, Qi J, et al. Value of circulating cell-free DNA in diagnosis of hepatocelluar carcinoma. World J Gastroenterol 2013;19:3143-9.
115. Ren N, Ye QH, Qin LX, Zhang BH, Liu YK, et al. Circulating DNA level is negatively associated with the long-term survival of hepatocellular carcinoma patients. World J Gastroenterol 2006;12:3911-4.
116. Iyer P, Zekri AR, Hung CW, Schiefelbein E, Ismail K, et al. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol 2010;88:107-11.
117. Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, et al. Plasma mSEPT9: A Novel Circulating Cell-free DNA-Based Epigenetic Biomarker to Diagnose Hepatocellular Carcinoma. EBioMedicine 2018;30:138-47.
118. Hu N, Fan XP, Fan YC, Chen LY, Qiao CY, et al. Hypomethylated Ubiquitin-Conjugating Enzyme2 Q1 (UBE2Q1) Gene Promoter in the Serum Is a Promising Biomarker for Hepatitis B Virus-Associated Hepatocellular Carcinoma. Tohoku J Exp Med 2017;242:93-100.
119. Jiao J, Watt GP, Stevenson HL, Calderone TL, Fisher-Hoch SP, et al. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: Prevalence and risk factors. Hepatol Commun 2018;2:718-31.
120. Liao W, Yang H, Xu H, Wang Y, Ge P, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016;7:40481-90.
121. Garcia-Fernandez N, Macher HC, Rubio A, Jimenez-Arriscado P, Bernal-Bellido C, et al. Detection of p53 Mutations in Circulating DNA of Transplanted Hepatocellular Carcinoma Patients as a Biomarker of Tumor Recurrence. Adv Exp Med Biol 2016;924:25-8.
122. Xu H, Zhu X, Xu Z, Hu Y, Bo S, et al. Non-invasive Analysis of Genomic Copy Number Variation in Patients with Hepatocellular Carcinoma by Next Generation DNA Sequencing. J Cancer 2015;6:247-53.
123. Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24.
124. Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open 2012;2:e000825.
125. Zhu HT, Hasan AM, Liu RB, Zhang ZC, Zhang X, et al. Serum microRNA profiles as prognostic biomarkers for HBV-positive hepatocellular carcinoma. Oncotarget 2016;7:45637-48.
126. Fu Y, Wei X, Tang C, Li J, Liu R, et al. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol Lett 2013;6:1811-5.
127. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med 2015;47:e184.
128. Huang JT, Liu SM, Ma H, Yang Y, Zhang X, et al. Systematic Review and Meta-Analysis: Circulating miRNAs for Diagnosis of Hepatocellular Carcinoma. J Cell Physiol 2016;231:328-35.
129. Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH, et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer 2016;138:714-20.
130. Luo J, Chen M, Huang H, Yuan T, Zhang M, et al. Circulating microRNA-122a as a diagnostic marker for hepatocellular carcinoma. Onco Targets Ther 2013;6:577-83.
131. Fang T, Lv H, Lv G, Li T, Wang C, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun 2018;9:191.
132. Zhang J, Lin H, Wang XY, Zhang DQ, Chen JX, et al. Predictive value of microRNA-143 in evaluating the prognosis of patients with hepatocellular carcinoma. Cancer Biomark 2017;19:257-62.
133. Zhao Q, Sun X, Liu C, Li T, Cui J, et al. Expression of the microRNA-143/145 cluster is decreased in hepatitis B virus-associated hepatocellular carcinoma and may serve as a biomarker for tumorigenesis in patients with chronic hepatitis B. Oncol Lett 2018;15:6115-22.
134. Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 2011;45:355-60.
135. Chen L, Chu F, Cao Y, Shao J, Wang F. Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumour Biol 2015;36:7439-47.
136. Amr KS, Ezzat WM, Elhosary YA, Hegazy AE, Fahim HH, et al. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene 2016;575:66-70.
137. Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One 2011;6:e26971.
138. Li F, Wang F, Zhu C, Wei Q, Zhang T, et al. miR-221 suppression through nanoparticle-based miRNA delivery system for hepatocellular carcinoma therapy and its diagnosis as a potential biomarker. Int J Nanomedicine 2018;13:2295-307.
139. Bhattacharya S, Steele R, Shrivastava S, Chakraborty S, Di Bisceglie AM, et al. Serum miR-30e and miR-223 as novel noninvasive biomarkers for hepatocellular carcinoma. Am J Pathol 2016;186:242-7.
140. Okajima W, Komatsu S, Ichikawa D, Miyamae M, Kawaguchi T, et al. Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget 2016;7:53820-36.
141. Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol 2015;16:804-15.
142. Gui J, Tian Y, Wen X, Zhang W, Zhang P, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183-93.
143. Chen Y, Dong X, Yu D, Wang X. Serum miR-96 is a promising biomarker for hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Int J Clin Exp Med 2015;8:18462-8.
144. Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet 2015;11:e1004873.
145. Xu Y, Bu X, Dai C, Shang C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour Biol 2015;36:4773-6.
146. Zhuang LP, Meng ZQ. Serum miR-224 reflects stage of hepatocellular carcinoma and predicts survival. Biomed Res Int 2015;2015:731781.
147. Cho HJ, Kim JK, Nam JS, Wang HJ, Lee JH, et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem 2015;48:1073-8.
148. Kim SS, Cho HJ, Nam JS, Kim HJ, Kang DR, et al. Plasma MicroRNA-21, 26a, and 29a-3p as predictive markers for treatment response following transarterial chemoembolization in patients with hepatocellular carcinoma. J Korean Med Sci 2018;33:e6.
149. Cho HJ, Kim SS, Nam JS, Kim JK, Lee JH, et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol 2017;41:181-9.
150. Xiang ZL, Zhao XM, Zhang L, Yang P, Fan J, et al. MicroRNA-34a expression levels in serum and intratumoral tissue can predict bone metastasis in patients with hepatocellular carcinoma. Oncotarget 2016;7:87246-56.
151. Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer 2015;112:532-8.
152. Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, et al. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids 2015;4:e233.
153. El-Tawdi AH, Matboli M, Shehata HH, Tash F, El-Khazragy N, et al. Evaluation of circulatory RNA-based biomarker panel in hepatocellular carcinoma. Mol Diagn Ther 2016;20:265-77.
154. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int 2013;2013:136106.
155. El-Tawdi AH, Matboli M, El-Nakeep S, Azazy AE, Abdel-Rahman O. Association of long noncoding RNA and c-JUN expression in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol 2016;10:869-77.
156. Tang J, Zhuo H, Zhang X, Jiang R, Ji J, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis 2014;5:e1549.
157. Wang X, Zhang W, Tang J, Huang R, Li J, et al. LINC01225 promotes occurrence and metastasis of hepatocellular carcinoma in an epidermal growth factor receptor-dependent pathway. Cell Death Dis 2016;7:e2130.
158. Tang J, Jiang R, Deng L, Zhang X, Wang K, et al. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget 2015;6:4505-15.
159. Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci 2016;107:149-54.
160. Jing W, Gao S, Zhu M, Luo P, Jing X, et al. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncol Rep 2016;36:1085-92.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Guerriero P, Moshiri F, Lupini L, Sabbioni S, Negrini M, Callegari E. Circulating tumor DNAs and non-coding RNAs as potential biomarkers for hepatocellular carcinoma diagnosis, prognosis and response to therapy. Hepatoma Res 2019;5:6. http://dx.doi.org/10.20517/2394-5079.2018.108
AMA Style
Guerriero P, Moshiri F, Lupini L, Sabbioni S, Negrini M, Callegari E. Circulating tumor DNAs and non-coding RNAs as potential biomarkers for hepatocellular carcinoma diagnosis, prognosis and response to therapy. Hepatoma Research. 2019; 5: 6. http://dx.doi.org/10.20517/2394-5079.2018.108
Chicago/Turabian Style
Guerriero, Paola, Farzaneh Moshiri, Laura Lupini, Silvia Sabbioni, Massimo Negrini, Elisa Callegari. 2019. "Circulating tumor DNAs and non-coding RNAs as potential biomarkers for hepatocellular carcinoma diagnosis, prognosis and response to therapy" Hepatoma Research. 5: 6. http://dx.doi.org/10.20517/2394-5079.2018.108
ACS Style
Guerriero, P.; Moshiri F.; Lupini L.; Sabbioni S.; Negrini M.; Callegari E. Circulating tumor DNAs and non-coding RNAs as potential biomarkers for hepatocellular carcinoma diagnosis, prognosis and response to therapy. Hepatoma. Res. 2019, 5, 6. http://dx.doi.org/10.20517/2394-5079.2018.108
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 9 clicks
Cite This Article 9 clicks



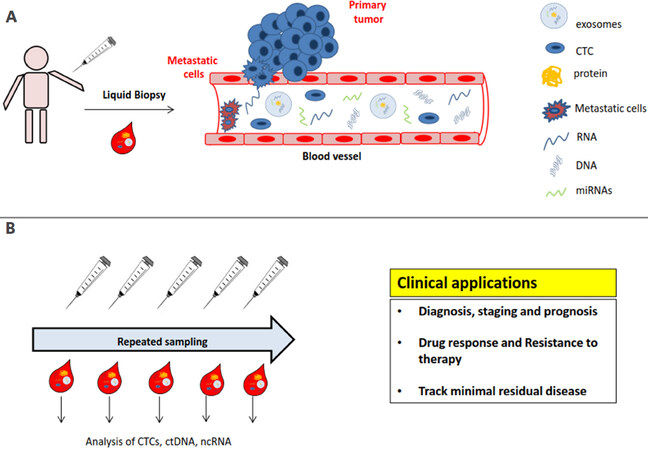









Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.