Artificial intelligence and its role in guiding liver-directed therapy for hepatocellular carcinoma: Is it ready for prime time?
Abstract
Artificial intelligence (AI) is an innovative discipline in medicine, impacting both hepatology and hepato-pancreato-biliary surgery, ensuring reliable outcomes because of its repeatable and efficient algorithms. A considerable number of studies about the efficiency of AI in the management of hepatocellular carcinoma (HCC) have been published. While its diagnostic role is well recognized, providing large amounts of quantitative radiological HCC features, its use in HCC treatment is still debated. Innovative use of AI may help to select the best approach for each patient as it is able to predict the outcomes after resection and/or other treatments. In this review, we assess the role of AI in selecting the best therapeutic option and predicting long-term risks after surgical or interventional treatments for HCC patients. Further studies are needed to consolidate AI applications.
Keywords
INTRODUCTION
The concept of artificial intelligence (AI) was developed as early as the 1950s[1], though it gained prominence decades later. AI is defined as a branch of informatics engaged in the design of software capable of providing performance to solve real-world problems, whose results must be equal to or better than human ones. It applies to several fields, including medicine[2,3].
Machine learning (ML) is part of AI in which computer programs learn from data, improving with experience. It is already a validated medical technique based on handcrafted features, while deep learning (DL) is a more recent subfield of ML[4].
In ML, clinicians and researchers list quantitative visual features and put them in a classification algorithm that allows them to categorize data[5]. Random Forest (RF) and support vector machines (SVM) are among the most used ML models.
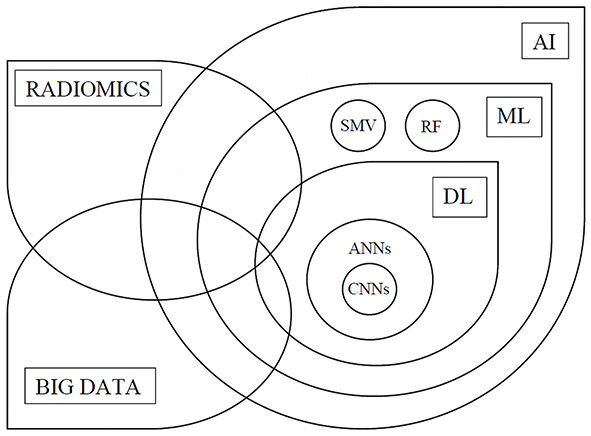
On the other hand, DL can be considered as a more complex branch of ML that requires specific hardware and usually analyzes more parameters than the basic ML. DL was designed with human neuroanatomy in mind, and it is organized in multiple layers (from input to output data, with more intermediate layers in between)[6]. Artificial neural networks (ANNs) are the principal branch of DL, with convolutional neural networks (CNNs) being a subset of ANNs algorithm [Figure 1][7,8]. A neural network is divided into three levels (layers): input layer, output layer, and hidden layer. The input layer comprises the data available for the analysis and the outcome layers comprise the outcome. The network learns how to associate an input with a specific output through the utilization of examples. Once the “network training” is completed, it can link new outputs to other inputs not used during the previous phase[9]. DL is not a handcrafted system. It automatically finds features associated with a particular outcome [Table 1].
Figure 1. Relationship between artificial intelligence, machine learning, deep learning, and radiomics. AI: Artificial intelligence;
A schematization of the difference between artificial intelligence, machine learning, and deep learning
| Artificial intelligence | Machine learning | Deep learning | |
| Definition | Branch of informatics engaged in designing software capable of providing performance by imitating human thinking and action | Subset of AI that automates analytical model building | Subset of ML based on artificial neural networks |
| Application | Building programs that utilize software to solve problems | Providing systems with the ability to automatically learn and improve performance from experience and data utilization | Providing systems with the ability to automatically process data and create patterns, improving performance via neural networks |
The use of AI methodologies in healthcare research increased during the last decade as a result of the advantageous ability to receive, process and interpret the complex mathematical elaboration of a large amount of data using only computer algorithms[8].
Notably, AI has proven its efficacy in skin cancer management, diagnosis of diabetic retinopathy and eye disease, identification of prostate cancer and brain metastasis, polyp detection during colonoscopy, and fractures identification on X-Ray[10-15].
HCC represents the fifth most widespread primitive cancer and one of the principal causes of malignancy-related death in the world[16]. HCC diagnosis is based on its characteristic radiological features in association with an α-fetoprotein (AFP) increase, and does not generally require a liver biopsy[17]. The radiological criteria widely known as LI-RADS (Liver Imaging Reporting and Data System) achieved good reliability by reducing imaging interpretation, facilitating communication with referring physicians, and improving research quality[18,19].
The cornerstone of treatment selection for HCC patients is the stage of the disease. For instance, liver transplantation (LT), liver resection (LR), or radiofrequency ablation (RFA) are all recommended in very-early and early-stage HCC[20]. In this context, identifying novel approaches to predict outcomes for each specific treatment among HCC patients is of paramount importance[6].
ARTIFICIAL INTELLIGENCE, RADIOMICS AND HEPATOCELLULAR CARCINOMA
Radiomics has rapidly emerged as a personalized medicine technology due to its ability to extract quantitative data from radiological images that cannot be detected by the human eye. It provides crucial information that can be processed, reducing errors, and optimizing timing in a diagnostic-therapeutic setting[21]. Its development is based on technological improvements and AI[8]. Radiomics generally includes several steps in its workflow:
- acquisition of images,
- selection of the region of interest (segmentation), which defines the volume for the features extraction phase,
- elaboration of features by software with pre- or post-processing tools (exploratory analysis),
- creation of a statistical model to select main data capable of predicting the objective of the analysis (model building)[22].
Each phase presents some criticalities and sources of variability that can affect the validity of the final model. Nevertheless, repeatable and efficient algorithms ensure reliable outcomes because of AI[23].
The refinement of prognostic models is fundamental to avoid unnecessary procedures and facilitate the individual management of HCC patients[24]. In the era of personalized medicine, the innovative use of AI might analyze certain tumor characteristics to predict the risk for recurrence or survival after resection, optimizing their effects with standardized risk stratification models[25].
LIVER TRANSPLANTATION
LT is the principal curative therapy for early-stage HCC[17]. Unfortunately, not all patients can benefit from it because the demand for LT exceeds the donor organ availability, and most of them undergo a subsidiary treatment, such as LR[26,27].
Linear regression and standard predictive modeling are fundamental for organ selection. Nowadays, donor risk index (DRI), model for end-stage liver disease (MELD), survival outcome following LT (SOFT), and the balance of risk score (BAR) are widely accepted models[28-31]. However, AI models are showing great potential in predicting LT short and long-term outcomes.
ML classifiers can be used to: list potential liver recipients and eventually remove them from the waiting list, and help to match and prognosticate candidates who may benefit from LT[32]. Moreover, they may identify factors that influence recurrence and survival and increase the use of marginal organs[32,33].
In terms of prediction, ANNs are the most popular classifiers due to their excellent capacity to analyze the numerous data involved in LT, including donor and recipient variables[34].
Most articles available focus on the risk of liver failure and overall survival at 3 months and/or at 1 year[34,35]. Briceño et al. explained that many of the current predictive models (i.e., MELD, DRI, SOFT, BAR) are too simplistic, because they assume a linearity among survival and LT variables, pointing out that LT follows a nonlinear pattern[35]. For this reason, ML scoring systems, using ANNs in conjunction with clinical judgment, represent the best way to predict the probability of graft survival and liver failure. ANNs allowed the authors to predict 3-month outcomes, achieving an area under the curve (AUC) of 0.82 and an accuracy of 0.91.
Some articles examined graft survival in the very short term, considering survival predictors at less than 30 days. Lau et al. elaborated an ANNs model to encourage clinical decisions, specifically for marginal organ allocation[36]. They identified the top 15 donor and recipient characteristics from a dataset of 276 variables, obtaining an AUC of 0.84, which is higher compared to traditional scoring systems.
Liu et al. focused on the prediction of patient survival after LT and created a ML model which includes patient’s blood tests within 9 days before surgery[37]. Their experimental model outperformed the MELD score in predicting postoperative survival, achieving a specificity of 0.815 and an AUC of 0.771.
AI does not yet have a widely recognized role in LT. However, because of the lack of deceased donor organs, it is crucial to acquire and standardize a method to help clinicians to maximize LT. In this context, a recent multicenter Korean study[38] developed and validated a novel AI model, MoRAL-AI, to predict cancer recurrence after LT. The study involved 563 HCC patients from three principal LT centers in Korea who underwent LT. Some of the most influencing factors in MoRAL-AI were tumor diameter and number, AFP, age, and portal invasion. The MoRAL-AI demonstrated a C-index of 0.75, showing a significantly better capacity for predicting cancer recurrence after LT compared to standard predictive models.
Similarly, Ivanics et al. gathered data from 739 transplanted patients, including preoperative imaging, locoregional therapy and response, and post-transplantation outcomes, to elaborate an AI model with the purpose of predicting HCC recurrence with excellent results[39].
According to ClinicalTrials.gov (https://clinicaltrials.gov/), we are waiting for the final results of a study (NCT05200195) by Lai et al., which includes more than 4000 transplanted HCC patients during the period 2000-2018 (international cohort, 3670 patients across 17 centers in Europe and Asia; external validation cohort, 356 patients transplanted at the Columbia University, New York). This large international study would prove that AI might be widely used in clinical settings in the near future.
The review of the literature is summarized in Table 2.
A summary of the articles describing the role of artificial intelligence in the prediction of hepatocellular carcinoma recurrence and/or survival after liver transplantation
| Author | Year | Country | Method | Aim | Findings |
| Kazemi et al.[33] | 2019 | Iran | ML | To identify effective factors for patient survival after LT | The model predicted survival based on 26 identified effective factors. The AUC and model sensitivity were 0.90 and 0.81 |
| Briceño et al.[35] | 2014 | Spain | ANNs | To use ANNs for D-R matching in LT and to compare its accuracy with validated scores | The models showed their best performance in predicting the probability of graft-survival (90.79%) and -loss (71.42%) for each D-R pair |
| Lau et al.[36] | 2017 | Australia | ML | To predict graft failure using donor and recipient factors, based on local data sets | The combination of factors used in DRI with the MELD score obtains an AUC of 0.764, whereas survival outcomes after LT score obtain an AUC of 0.638 |
| Liu et al.[37] | 2020 | Taiwan | ML | To predict 30-day postoperative survival based on patient's preoperative physiological measurement values | The model achieves an AUC of 0.771 and a specificity of 0.815, showing superior discrimination power in predicting postoperative survival |
| Nam et al.[38] | 2020 | Korea | AI | To develop a novel model to predict tumor recurrence after LT by adopting AI (MoRAL-AI) | The largest weighted parameter in the MoRAL-AI was tumor diameter, followed by AFP, age, and PIVKA-II. The MoRAL-AI had better predictability of tumor recurrence after LT than conventional models |
| Ivanics et al.[39] | 2022 | Canada | ML | To leverage ML to develop an accurate post-LT HCC recurrence prediction calculator, to optimize the utility of limited donor organs | The model performed well with a concordance of 0.72 (95%CI: 0.63-0.81) and was not significantly outperformed |
LIVER RESECTION
The European Association for the Study of the Liver (EASL) Clinical Practice Guidelines[40] recommends LR for patients with very-early/early-stage HCC (stage 0-A) and well-preserved liver function.
Although the increasing evolution of surgery and the accuracy of perioperative clinical monitoring, post-hepatectomy liver failure (PHLF) is one of the most alarming complications after LR, particularly in cirrhotic patients, and is associated with a high risk of postoperative mortality[41]. The accurate assessment of the volume of the future liver remnant (FRL) is pivotal to reducing PHLF. Despite the globally accepted role of contrast-enhanced tomography scan (CT) for FLR assessment, recent fully automated liver volume assessments are showing good speed and accuracy compared to manual segmentation[42,43].
Accurate knowledge of liver anatomy is critical for any successful LR to reduce the risk of PHLF. In several cases, liver anatomy is so complex that it is difficult to reconstruct it mentally during LR, and preoperative imaging might be insufficient. Nowadays, FLR may be estimated with better accuracy, and HCC resectability may be carefully assessed thanks to innovative 3D visualization techniques[44]. However, 3D simulation techniques are costly and time-consuming. ML could be exploited to accelerate the 3D modelling process through quicker image acquisition and segmentation[45].
Several scoring systems are employed in clinical practice to determine the quality of the liver and predict PHLF, including Child-Pugh grade, MELD, albumin-bilirubin (ALBI), platelet-albumin-bilirubin (PALBI), the aspartate aminotransferase to platelet ratio index (APRI) and the fibrosis index (FIB-4)[29,46-50]. However, most of these scoring systems are not always reliable for PHLF prediction. In this context, Mai et al. developed and validated an ANNs model in which they included blood tests (platelet count, total bilirubin, prothrombin time, and aspartate aminotransferase) and expected FLR[50]. This study showed a significantly more accurate prediction of severe PHLF (AUC value of 0.880) compared to the predictive systems.
Despite significant progress in the management of HCC, the recurrence rate after LR is still high (70%-80%) and is associated with a poor prognosis[51,52]. At the moment, there are no reliable risk stratification models for HCC recurrence after LR.
Several clinical features including number of nodules, maximum nodule diameter, cytological differentiation, and microvascular invasion (MVI) have been used to predict HCC recurrence, using a conventional Cox proportional hazards regression model[53-56], without satisfactory results.
Biomarkers, including gene mutational status, proteins, and micro-RNA (miRNA), have been shown to be better survival predictors than TNM staging[57,58].
MVI represents an independent predictive factor of recurrence after LR and is consequently associated with poor prognosis[59]. Radiomic signatures have been elaborated, focusing on preoperative MVI detected on CT or magnetic resonance imaging (MRI), to enable HCC recurrence prediction[60-62]. Moreover,
Ho et al. explored the 1-, 3- and 5-year disease-free survival (DFS) prediction in HCC patients undergoing LR using ANNs[64]. The ANNs model collected demographic data, clinical features, surgical details, and outcomes, outperforming the conventional logistic regression models for prediction accuracy.
Saillard et al. elaborated a survival prediction model for HCC patients who underwent LR[65]. They processed all the HCC digital slides to create a DL model, achieving higher accuracy compared to main scores that include clinical, biological, and pathological features.
Schoenberg et al. recruited 181 patients who underwent partial LR and used their data to develop a ML algorithm to predict DFS after LR[26]. This model was built on 26 preoperatively laboratory variables and standard clinical-pathological parameters. A RF-based workflow was employed to process data, reporting 6 relevant DFS predictors: modified Glasgow Prognostic Score (mGPS), partial thromboplastin time (aPTT), C-reactive protein (CRP), age at surgery, largest tumor size and number of lesions.
Ji et al. acquired data from 470 patients to elaborate preoperative and postoperative ML-based radiomics models to predict HCC recurrence after LR[66]. The available preoperative CT imaging, AFP value, ALBI grade and liver cirrhosis were used to create the preoperative model. The postoperative model was created by adding the presence of satellite nodules to the other predictors. The two models presented a C-index of 0.733 and 0.801, respectively, showing a superior accuracy after comparison with other non-radiomic models.
Zheng et al. tried to predict both HCC recurrence and postoperative survival in patients with solitary HCC, who were selected for LR using a ML-based radiomics model[67]. This model reached a good predictive survival accuracy with a C-index of 0.71. Moreover, adding TNM and Barcelona Clinic Liver Cancer (BCLC) stage to this model, they demonstrated a further increase of C-index.
Similarly, Zhang et al. used a radiomics model based on dynamic contrast-enhanced MRI to predict the survival of patients who underwent LR for HCC[62]. The tumor, perilesional, and non-tumoral parenchyma imaging was used to obtain three radiomics scores. The non-tumoral parenchyma score had the highest prognostic role showing a C-index of 0.72.
As already discussed, most of the models are based on preoperative imaging. Otherwise, Shen et al. proposed a ML-radiomics model trained on CT imaging within one month after treatment (surgery or ablation)[68]. They analyzed lesions suspicious for HCC recurrence. The AUC, accuracy, specificity, and sensitivity of this model for detecting recurrence were 0.89, 0.86, 0.75 and 0.91, respectively.
According to ClinicalTrials.gov (https://clinicaltrials.gov/), Zhujiang et al. are employing the innovative Watson computer system (an AI platform developed by IBM®) to select the best treatment options for each patient affected by HCC. This innovative ML-based system acquires a large amount of structured and unstructured data and selects the main clinical features to guide surgical decisions. A more comprehensive and reliable ML-based decision model available in daily clinical practice should be created. In this context, Zhejiang et al.’s study might be crucial.
The review of the literature is summarized in Table 3.
A summary of the articles describing the role of artificial intelligence in the prediction of hepatocellular carcinoma recurrence and/or survival after liver resection
| Author | Year | Country | Method | Aim | Findings |
| Schoenberg et al.[26] | 2020 | Germany | ML | To develop a ML algorithm to predict which patients are sufficiently treated with LR | Six predictors identified: mGPS, aPTT, CRP, largest tumor size, number of lesions and age at the time of LR. Predictive value of the model 0.78 |
| Mai et al.[50] | 2020 | China | ANNs | To establish and validate an ANNs model to predict severe PHLF | ANNs accurately predicted the risk of severe PHLF after hemihepatectomy |
| Ma et al.[60] | 2019 | China | Radiomics | To develop and validate a radiomics nomogram for preoperative prediction of MVI | The proposed radiomics nomogram can preoperatively predict MVI |
| Xu et al.[61] | 2019 | China | Radiomics | To develop radiomic features from CT to predict MVI and clinical outcomes | Good performance for predicting MVI and clinical outcomes, without providing statistically significant added value to radiographic scores |
| Zhang et al.[62] | 2020 | China | Radiomics | To evaluate the efficiency of gadoxetic acid-enhanced MRI-based radiomics features for prediction of OS after LR | The combined model incorporating clinic-radiological and radiomics features showed an improved predictive performance with C-index of 0.92 |
| Dong et al.[63] | 2020 | China | Radiomics | To establish a radiomic algorithm based on grayscale US images for preoperative predictions of MVI | Potential value to facilitate preoperative prediction of MVI |
| Ho et al.[64] | 2012 | Taiwan | ANNs | To develop prediction models for 1-, 3- and 5-year DFS after LR | The ANNs model outperformed the LR and DT models for predicting disease-free survival |
| Saillard et al.[65] | 2020 | France | DL | To build models for predicting survival after LR | Models had a higher discriminatory power than a score combining all baseline variables and helped predict prognosis |
| Ji et al.[66] | 2019 | China | ML; radiomics | To explore the potential of radiomics coupled with ML algorithms to improve the predictive accuracy for HCC recurrence | The two models showed superior prognostic performance, with C-index of 0.733-0.801 compared with models without radiomics |
| Zheng et al.[67] | 2018 | China | Radiomics | To estimate postoperative recurrence and survival in patients with solitary HCC | The model had a better prognostic performance as compared with traditional staging systems |
| Shen et al.[68] | 2021 | China | Radiomics | To develop a radiomics algorithm, improving the performance of detecting recurrence, based on post-treatment CT images within one month | The algorithm integrated radiomic features of post-treatment CT and showed superior performance compared with the conventional AFP |
RADIOFREQUENCY ABLATION
According to EASL[40], RFA is suggested for patients affected by HCC at very-early stage and early stage (BCLC stage 0 and A) with sufficient functional liver reserve[20]. RFA can also be considered as an effective bridging therapy to LT for cirrhotic patients with small HCCs, and a valuable treatment option for patients with unresectable HCfC[22,69].
HCC recurrence after RFA is currently still frequent. DFS at 5 years is more than 70% and this is due to concomitant risk factors, including underlying viral hepatitis and cirrhosis[70-72]. Therefore, a ML-based predictive model for HCC recurrence is needed for HCC patients who undergo RFA to support clinical decision-making. In this context, patients who are identified as high-risk subjects could be selected for a tight follow-up.
Wu et al. developed an ANNs model to predict 1- and 2-year DFS in HCC patients scheduled for RFA[73]. They found that this model, which includes 15 clinical variables, had a better prediction power than scores with limited inputs, reaching an AUC of 0.77 and 0.72 for 1-year and 2-year DFS, respectively.
Liang et al. elaborated a predictive model for HCC recurrence focusing on patients who underwent RFA as their first treatment[74]. In this study, they tried to select few significant clinical features from a total of 16. They employed 5 widely used selection methods and combined them with SVM to develop a predictive model with better sensitivity and specificity.
Currently, the research, development, and clinical application of innovative post-RFA predictive models remain inadequate.
The review of the literature is summarized in Table 4.
A summary of the articles describing the role of artificial intelligence in the prediction of hepatocellular carcinoma recurrence and/or survival after radiofrequency ablation and transcatheter arterial chemoembolization
| Author | Year | Country | Method | Aim | Findings |
| Wu et al.[73] | 2017 | Taiwan | ANNs | To build ANNs models with HCC-related variables to predict the 1- and 2-year DFS after RFA | Parameters for performance assessment of 1-year DFS prediction were: accuracy 85.0%, sensitivity 75.0%, specificity 87.5%, and an AUC 0.84. For 2-year DFS prediction, the values of accuracy, sensitivity, specificity, and AUC were 67.9%, 50.0%, 85.7%, and 0.75, respectively |
| Liang et al.[74] | 2014 | Taiwan | ML | To develop recurrence predictive models after RFA | The developed model had a sensitivity, specificity, accuracy, PPV, NPV, and AUC of 67%, 86%, 82%, 69%, 90%, and 0.69, respectively |
| Mähringer-Kunz et al.[81] | 2020 | Germany | ANNs | To develop a survival prediction model for patients undergoing TACE using novel ML algorithms and to compare it to conventional prediction scores | The model had a promising performance for predicting 1-year survival, with an AUC of 0.77 ± 0.13, a PPV of 87.5%, and a NPV of 68.0% |
| Liu et al.[82] | 2020 | China | Radiomics | To validate an AI-based radiomics strategy for predicting responses to a first TACE treatment by quantitatively analyzing CEUS | The AUC for the model was 0.93. It can be used to achieve accurate and personalized prediction |
| Oezdemi et al.[83] | 2020 | USA | ML | To predict the response to TACE using CEUS-derived imaging features | The model may help to provide personalized therapy planning. Number of vessels, number of bifurcations and vessel-to-tissue ratio are the dominant features for the prediction of long-term TACE response |
| Morshid et al.[84] | 2019 | USA | ML | To evaluate a fully automated ML algorithm that uses pre-therapeutic quantitative CT imaging features and clinical factors to predict response to TACE | The model’s response prediction accuracy rate was 74.2%, using a combination of the BCLC stage plus quantitative imaging features vs. 62.9% using the BCLC stage alone |
| Peng et al.[85] | 2020 | China | ANNs | To train and validate a model of DL for the prediction of the response of patients with intermediate-stage HCC undergoing TACE | The model had an accuracy of 84.3% and AUC of 0.97, 0.96, 0.95, and 0.96 for complete response, partial response, and stable disease, respectively |
| Abajian et al.[86] | 2018 | USA | ML | To develop a method to predict response to TACE treatment prior to LR | The model predicted TACE response with an overall accuracy of 78% |
TRANSCATHETER ARTERIAL CHEMOEMBOLIZATION
HCC receives its blood supply primarily from hepatic artery branches. Hence, transcatheter arterial chemoembolization (TACE) represents a highly selective therapeutic option that might deliver a high concentration of chemotherapeutics to target tumors, sparing the surrounding hepatic tissue[75].
According to the BCLC classification, ACE is the gold standard for cirrhotic patients with multinodular, unresectable HCC (stage B) with preserved liver function[22,24].
One of the best outcome predictors for cirrhotic patients affected by stage B HCC is the response to the first TACE. Large tumors often respond unsatisfactorily to a single TACE session. Repeating sessions increase the risk of multiple adverse effects including liver and/or kidney failure, gastroduodenal ulceration, and death[76,77].
In this context, several conventional scoring systems have been created to support critical clinical decisions in case of uncertain retreatment with TACE, including Assessment for Retreatment with TACE (ART), Child-Pugh, Objective Radiological Response (SNACOR), and Child-Pugh and Response (ABCR)[78-80].
Mähringer-Kunz et al. developed an ANNs prediction model for survival after TACE combining all of the above-mentioned scores[81]. They elaborated a 1-year survival model obtaining better results compared to ART, SNACOR, and ABCR scores.
Liu et al. employed radiomics to predict personalized response to a first TACE session by quantitatively analyzing contrast-enhanced ultrasound (CEUS) cines[82]. This DL radiomics-based CEUS model established high reproducibility and presented an AUC of 0.93.
Oezdemir et al. used morphological and anatomical features detected by CEUS to create a ML-based predictive model for HCC patients treated with TACE[83]. Their study involved 36 HCC patients and revealed that number of vessels, number of bifurcations, and vessel-to-tissue ratio represented the principal features for the prediction of long-term TACE response, with an accuracy of 86%.
Morshid et al. managed to improve the accuracy of predicting HCC response to TACE thanks to a ML-based model, which combines BCLC stage and quantitative image features[84]. They achieved a prediction accuracy rate of 74.2%.
Peng et al. used preoperative CT images to elaborate a DL prediction model of response to TACE[85]. Notably, they focused on every possible occurrence after TACE: complete response, partial response, stable disease, and progressive disease. This DL model reached an accuracy of 84.3% and an AUC tending to 1 for each occurrence after TACE.
Abajian et al. used ML to combine MRI images with clinical data to predict response to TACE in a case series of 36 patients, reaching an overall accuracy of 78%[86].
Hypoxia is the main mechanism of cell death induction in embolization because it produces complex molecular changes, promoting cell death or cell survival depending on genetic features[87]. Recent studies used AI to focus on the efficacy of TACE by analyzing these genetic characteristics. For example, Ziv et al. developed the linear SVM (a ML supervised model) to study genetic mutations[88]. They managed to identify the upregulation of the WNT/β-catenin signaling pathway, which is highly sensitive to embolization.
The review of the literature is summarized in Table 4.
MEDICAL THERAPY
In the era of personalized medicine, the knowledge of HCC molecular features (i.e., genome, transcriptome, proteome, and metabolome) can help stratify tumor aggressiveness and choose the best therapy accordingly. Notably, the identification of commonly altered pathways has traditionally been more difficult in HCC than in other cancers. In this context, AI can be helpful in the fields of target-based therapeutic discovery, systems-based approaches, and immune HCC landscape definition[89].
The targeted therapy with Sorafenib is widely used as first-line therapy in advanced HCC patients, but it is associated with delicate dose-limiting toxicity and a high risk of drug resistance[90-92]. In this context, AI should emerge as a tool to individually predict the risk for adverse effects in this group of patients. Abuhelwa et al. developed a predictive model including bilirubin, hemoglobin, and sex to define subgroups at risk of developing severe hand-foot syndrome[93]. Their model was validated with ML.
The progress made in immunotherapy (IT) with checkpoint inhibitors plays an increasingly decisive role in controlling the progression of HCC[94]. AI represents an emerging tool to validate predictive markers and anticipate the effectiveness of this innovative medical therapy.
Tumor immune microenvironment and cancer stem cells (CSCs) behavior are clearly related to HCC progression and drug resistance. ML has been recently applied to realize algorithms that highlight different stemness subtypes and predict response to IT according to the stemness index[95].
Sangro et al. reported how the expression of the inflammatory markers anti-programmed death 1 receptor (PD1) and PD-ligand 1 (PD-L1), and some particular immune gene signatures are associated with improved survival and response in patients treated with nivolumab, a PD1 inhibitor utilized in patients with advanced HCC[96]. In this setting, Zeng et al. trained CNNs using digital histological slides and gene signature status as labels and investigated three deep learning approaches to predict immune and inflammatory gene signatures directly from HCC histology[97]. The clustering-constrained attention multiple-instance learning (CLAM) model proved to be the best in predicting the upregulation of the immune gene signatures, with AUCs ranging from 0.78 to 0.91.
Lui et al. included 47 clinical variables in 6 different ML models to analyze the 1-year cancer-related mortality in advanced HCC patients treated with IT[98]. They demonstrated an optimal predictive capacity of ML compared to C-reactive protein and AFP in both the ImmunoTherapY score (CRAFITY) and ALBI score. In this study, it was also highlighted how baseline AFP, bilirubin, and alkaline phosphatase act as three common risk factors in all the models.
In the drug discovery setting, RF, SVM, neural networks, and the gradient boosting machine were compared by using nested cross-validation to evaluate the gene-disease association data from the Open Targets platform to predict therapeutic targets that are actively being pursued by pharmaceutical companies or are already on the market. On a test set, the neural network classifier achieved the best accuracy with an AUC of 0.76 in predicting novel targets[99].
Tong et al. employed the human protein-protein interaction (PPI) network to potentially map drug target genes working on a genetic dependency score[100]. They combined these manually collected drug target genes with gene expression profiles, and clinical data and they used SVM to build a drug target predictor. Similarly, Cetin-Atalay et al. used the CROssBAR innovative system to integrate a large number of biomedical data sources to define ligands of HCC-associated proteins as potential drug targets and repurpose known small molecule inhibitors as potential drug candidates[101]. They used ML as a virtual screening in target identification and drug discovery, developing DL-based drug/compound-target protein interaction predictors.
Table 5 summarizes the applications of AI in the treatment of HCC.
Artificial intelligence applications in the therapy of hepatocellular carcinoma
| Artificial intelligence and HCC treatment |
| Facilitates D-R matching for LT |
| Predicts short and long-term outcomes after LT |
| Predicts the risk of MVI after LR |
| Predicts PHLF after LR |
| Predicts RFS and OS after LR |
| Predicts RFS after RFA |
| Predicts response to TACE |
| Predicts medical therapy toxicity |
| Identifies markers associated with response to IT |
| Identifies new drug targets |
CONCLUSION
AI has become an important tool in HCC diagnosis, revealing a large amount of quantitative radiological HCC features. It has also shown very promising results in the fields of segmentation, prediction of recurrence and survival after specific treatments, thanks to its ability to process large quantities of data. It might become crucial in defining a tailored approach for each patient and may be routinely used in clinical practice for HCC management. Nevertheless, based on its complexity and the need for further validation, additional studies are necessary to implement its acceptance and diffusion. The most skeptical opponents to AI should consider that it is a tool to support human intelligence, not replace it.
DECLARATIONS
AcknowledgmentsWe thank Ariana M. Chirban, BS for reviewing the English.
Authors’ contributionsMade substantial contributions to conception and design of the study: Panettieri E, Campisi A
Made significant contributions to the writing of the manuscript: Campisi A, Bianchi V
Contributed to the production of the final draft and gave administrative, technical, and material support: Giuliante F, De Rose AM
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
2. Kaul V, Enslin S, Gross SA. History of artificial intelligence in medicine. Gastrointest Endosc 2020;92:807-12.
3. Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol 2019;25:1666-83.
4. Nam D, Chapiro J, Paradis V, Seraphin TP, Kather JN. Artificial intelligence in liver diseases: improving diagnostics, prognostics and response prediction. JHEP Rep 2022;4:100443.
5. Russell SJ, Norvig P. Artificial intelligence. A modern approach. New Jersey: Pearson Education, Inc.,; 2003.
6. Calderaro J, Seraphin TP, Luedde T, Simon TG. Artificial intelligence for the prevention and clinical management of hepatocellular carcinoma. J Hepatol 2022;76:1348-61.
7. Locke S, Bashall A, Al-adely S, Moore J, Wilson A, Kitchen GB. Natural language processing in medicine: a review. Trends Anaesth Crit Care 2021;38:4-9.
8. Feng B, Ma XH, Wang S, Cai W, Liu XB, Zhao XM. Application of artificial intelligence in preoperative imaging of hepatocellular carcinoma: current status and future perspectives. World J Gastroenterol 2021;27:5341-50.
9. Cucchetti A, Piscaglia F, Grigioni AD, et al. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: a pilot study. J Hepatol 2010;52:880-8.
10. Goyal M, Knackstedt T, Yan S, Hassanpour S. Artificial intelligence-based image classification methods for diagnosis of skin cancer: challenges and opportunities. Comput Biol Med 2020;127:104065.
11. Ting DSW, Cheung CY, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017;318:2211-23.
12. Cuocolo R, Cipullo MB, Stanzione A, et al. Machine learning for the identification of clinically significant prostate cancer on MRI: a meta-analysis. Eur Radiol 2020;30:6877-87.
13. Cho SJ, Sunwoo L, Baik SH, Bae YJ, Choi BS, Kim JH. Brain metastasis detection using machine learning: a systematic review and meta-analysis. Neuro Oncol 2021;23:214-25.
14. Barua I, Vinsard DG, Jodal HC, et al. Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta-analysis. Endoscopy 2021;53:277-84.
15. Hardalaç F, Uysal F, Peker O, et al. Fracture detection in wrist X-ray images using deep learning-based object detection models. Sensors 2022;22:1285.
16. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209-49.
17. Association For The Study Of The Liver, European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908-43.
18. Mitchell DG, Bruix J, Sherman M, Sirlin CB. LI-RADS (Liver Imaging Reporting and Data System): summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 2015;61:1056-65.
19. Liang Y, Xu F, Guo Y, et al. Diagnostic performance of LI-RADS for MRI and CT detection of HCC: a systematic review and diagnostic meta-analysis. Eur J Radiol 2021;134:109404.
20. Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 1999;19:329-38.
21. El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: Where are we? Hepatology 2014;60:1767-75.
22. Lewis S, Hectors S, Taouli B. Radiomics of hepatocellular carcinoma. Abdom Radiol 2021;46:111-23.
23. Castaldo A, De Lucia DR, Pontillo G, et al. State of the art in artificial intelligence and radiomics in hepatocellular carcinoma. Diagnostics 2021;11:1194.
25. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563-77.
26. Schoenberg MB, Bucher JN, Koch D, et al. A novel machine learning algorithm to predict disease free survival after resection of hepatocellular carcinoma. Ann Transl Med 2020;8:434.
27. Schoenberg MB, Bucher JN, Vater A, et al. Resection or transplant in early hepatocellular carcinoma. Dtsch Arztebl Int 2017;114:519-26.
28. Lozanovski VJ, Probst P, Arefidoust A, et al. Prognostic role of the donor risk index, the eurotransplant donor risk index, and the balance of risk score on graft loss after liver transplantation. Transpl Int 2021;34:778-800.
29. Kim WR, Mannalithara A, Heimbach JK, et al. MELD 3.0: the model for end-stage liver disease updated for the modern era. Gastroenterology 2021;161:1887-1895.e4.
30. Rana A, Jie T, Porubsky M, et al. The survival outcomes following liver transplantation (SOFT) score: validation with contemporaneous data and stratification of high-risk cohorts. Clin Transplant 2013;27:627-32.
31. Veerankutty FH, Jayan G, Yadav MK, et al. Artificial Intelligence in hepatology, liver surgery and transplantation: emerging applications and frontiers of research. World J Hepatol 2021;13:1977-90.
32. Khorsandi SE, Hardgrave HJ, Osborn T, et al. Artificial intelligence in liver transplantation. Transplant Proc 2021;53:2939-44.
33. Kazemi A, Kazemi K, Sami A, Sharifian R. Identifying factors that affect patient survival after orthotopic liver transplant using machine-learning techniques. Exp Clin Transplant 2019;17:775-83.
34. Wingfield LR, Ceresa C, Thorogood S, Fleuriot J, Knight S. Using artificial intelligence for predicting survival of individual grafts in liver transplantation: a systematic review. Liver Transpl 2020;26:922-34.
35. Briceño J, Cruz-Ramírez M, Prieto M, et al. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: results from a multicenter Spanish study. J Hepatol 2014;61:1020-8.
36. Lau L, Kankanige Y, Rubinstein B, et al. Machine-learning algorithms predict graft failure after liver transplantation. Transplantation 2017;101:e125-32.
37. Liu CL, Soong RS, Lee WC, Jiang GW, Lin YC. Predicting short-term survival after liver transplantation using machine learning. Sci Rep 2020;10:5654.
38. Nam JY, Lee JH, Bae J, et al. Novel model to predict HCC recurrence after liver transplantation obtained using deep learning: a multicenter study. Cancers 2020;12:2791.
39. Ivanics T, Nelson W, Patel MS, et al. The Toronto postliver transplantation hepatocellular carcinoma recurrence calculator: a machine learning approach. Liver Transpl 2022;28:593-602.
40. Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu, European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182-236.
41. Balzan S, Belghiti J, Farges O, et al. The “50-50 criteria” on postoperative day 5: an accurate predictor of liver failure and death after hepatectomy. Ann Surg 2005;242:824-9.
42. Wang K, Mamidipalli A, Retson T, et al. Members of the NASH Clinical Research Network. Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol Artif Intell 2019;1:180022.
43. Winkel DJ, Weikert TJ, Breit HC, et al. Validation of a fully automated liver segmentation algorithm using multi-scale deep reinforcement learning and comparison versus manual segmentation. Eur J Radiol 2020;126:108918.
44. Mise Y, Hasegawa K, Satou S, et al. How has virtual hepatectomy changed the practice of liver surgery? Ann Surg 2018;268:127-33.
45. Huff TJ, Ludwig PE, Zuniga JM. The potential for machine learning algorithms to improve and reduce the cost of 3-dimensional printing for surgical planning. Expert Rev Med Devices 2018;15:349-56.
46. Durand F, Valla D. Assessment of the prognosis of cirrhosis: child-pugh versus MELD. J Hepatol 2005;42 Suppl:S100-7.
47. Johnson PJ, Berhane S, Kagebayashi C, et al. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach-the ALBI grade. J Clin Oncol 2015;33:550-8.
48. Lu LH, Zhang YF, Mu-Yan C, et al. Platelet-albumin-bilirubin grade: Risk stratification of liver failure, prognosis after resection for hepatocellular carcinoma. Dig Liver Dis 2019;51:1430-7.
49. Zhou P, Chen B, Miao XY, et al. Comparison of FIB-4 index and child-pugh score in predicting the outcome of hepatic resection for hepatocellular carcinoma. J Gastrointest Surg 2020;24:823-31.
50. Mai RY, Lu HZ, Bai T, et al. Artificial neural network model for preoperative prediction of severe liver failure after hemihepatectomy in patients with hepatocellular carcinoma. Surgery 2020;168:643-52.
51. Kudo M. Systemic therapy for hepatocellular carcinoma: 2017 update. Oncology 2017;93 Suppl 1:135-46.
52. Llovet JM, Montal R, Villanueva A. Randomized trials and endpoints in advanced HCC: role of PFS as a surrogate of survival. J Hepatol 2019;70:1262-77.
53. D’Amico F, Schwartz M, Vitale A, et al. Predicting recurrence after liver transplantation in patients with hepatocellular carcinoma exceeding the up-to-seven criteria. Liver Transpl 2009;15:1278-87.
54. Ling Q, Liu J, Zhuo J, et al. Development of models to predict early post-transplant recurrence of hepatocellular carcinoma that also integrate the quality and characteristics of the liver graft: a national registry study in China. Surgery ;2018:155-64.
55. Agopian VG, Harlander-Locke M, Zarrinpar A, et al. A novel prognostic nomogram accurately predicts hepatocellular carcinoma recurrence after liver transplantation: analysis of 865 consecutive liver transplant recipients. J Am Coll Surg 2015;220:416-27.
56. Mehta N, Heimbach J, Harnois DM, et al. Validation of a risk estimation of tumor recurrence after transplant (RETREAT) score for hepatocellular carcinoma recurrence after liver transplant. JAMA Oncol 2017;3:493-500.
57. El-Gebaly F, Abou-Saif S, Elkadeem M, et al. Study of serum soluble programmed death ligand 1 as a prognostic factor in hepatocellular carcinoma in egyptian patients. Curr Cancer Drug Targets 2019;19:896-905.
58. Wang S, Zhang JH, Wang H, et al. A novel multidimensional signature predicts prognosis in hepatocellular carcinoma patients. J Cell Physiol 2019;234:11610-9.
59. Erstad DJ, Tanabe KK. Prognostic and therapeutic implications of microvascular invasion in hepatocellular carcinoma. Ann Surg Oncol 2019;26:1474-93.
60. Ma X, Wei J, Gu D, et al. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol 2019;29:3595-605.
61. Xu X, Zhang HL, Liu QP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol 2019;70:1133-44.
62. Zhang Z, Chen J, Jiang H, et al. Gadoxetic acid-enhanced MRI radiomics signature: prediction of clinical outcome in hepatocellular carcinoma after surgical resection. Ann Transl Med 2020;8:870.
63. Dong Y, Zhou L, Xia W, et al. Preoperative prediction of microvascular invasion in hepatocellular carcinoma: initial application of a radiomic algorithm based on grayscale ultrasound images. Front Oncol 2020;10:353.
64. Ho WH, Lee KT, Chen HY, Ho TW, Chiu HC. Disease-free survival after hepatic resection in hepatocellular carcinoma patients: a prediction approach using artificial neural network. PLoS One 2012;7:e29179.
65. Saillard C, Schmauch B, Laifa O, et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology 2020;72:2000-13.
66. Ji GW, Zhu FP, Xu Q, et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine 2019;50:156-65.
67. Zheng BH, Liu LZ, Zhang ZZ, et al. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer 2018;18:1148.
68. Shen JX, Zhou Q, Chen ZH, et al. Longitudinal radiomics algorithm of posttreatment computed tomography images for early detecting recurrence of hepatocellular carcinoma after resection or ablation. Transl Oncol 2021;14:100866.
69. Tiong L, Maddern GJ. Systematic review and meta-analysis of survival and disease recurrence after radiofrequency ablation for hepatocellular carcinoma. Br J Surg 2011;98:1210-24.
70. Hung HH, Chiou YY, Hsia CY, et al. Survival rates are comparable after radiofrequency ablation or surgery in patients with small hepatocellular carcinomas. Clin Gastroenterol Hepatol 2011;9:79-86.
71. Kao WY, Chiou YY, Hung HH, et al. Risk factors for long-term prognosis in hepatocellular carcinoma after radiofrequency ablation therapy: the clinical implication of aspartate aminotransferase-platelet ratio index. Eur J Gastroenterol Hepatol 2011;23:528-36.
72. Kao WY, Chiou YY, Hung HH, et al. Serum alpha-fetoprotein response can predict prognosis in hepatocellular carcinoma patients undergoing radiofrequency ablation therapy. Clin Radiol 2012;67:429-36.
73. Wu CF, Wu YJ, Liang PC, Wu CH, Peng SF, Chiu HW. Disease-free survival assessment by artificial neural networks for hepatocellular carcinoma patients after radiofrequency ablation. J Formos Med Assoc 2017;116:765-73.
74. Liang JD, Ping XO, Tseng YJ, Huang GT, Lai F, Yang PM. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Comput Methods Programs Biomed 2014;117:425-34.
75. Pesapane F, Nezami N, Patella F, Geschwind JF. New concepts in embolotherapy of HCC. Med Oncol 2017;34:58.
76. Kim BK, Kim SU, Kim KA, et al. Complete response at first chemoembolization is still the most robust predictor for favorable outcome in hepatocellular carcinoma. J Hepatol 2015;62:1304-10.
77. Marcacuzco Quinto A, Nutu OA, San Román Manso R, et al. Complications of transarterial chemoembolization (TACE) in the treatment of liver tumors. Cir Esp 2018;96:560-7.
78. Sieghart W, Hucke F, Pinter M, et al. The ART of decision making: retreatment with transarterial chemoembolization in patients with hepatocellular carcinoma. Hepatology 2013;57:2261-73.
79. Adhoute X, Penaranda G, Naude S, et al. Retreatment with TACE: the ABCR SCORE, an aid to the decision-making process. J Hepatol 2015;62:855-62.
80. Mähringer-Kunz A, Weinmann A, Schmidtmann I, et al. Validation of the SNACOR clinical scoring system after transarterial chemoembolisation in patients with hepatocellular carcinoma. BMC Cancer 2018;18:489.
81. Mähringer-Kunz A, Wagner F, Hahn F, et al. Predicting survival after transarterial chemoembolization for hepatocellular carcinoma using a neural network: a pilot study. Liver Int 2020;40:694-703.
82. Liu D, Liu F, Xie X, et al. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur Radiol 2020;30:2365-76.
83. Oezdemir I, Wessner CE, Shaw C, Eisenbrey JR, Hoyt K. Tumor vascular networks depicted in contrast-enhanced ultrasound images as a predictor for transarterial chemoembolization treatment response. Ultrasound Med Biol 2020;46:2276-86.
84. Morshid A, Elsayes KM, Khalaf AM, et al. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol Artif Intell 2019;1:e180021.
85. Peng J, Kang S, Ning Z, et al. Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging. Eur Radiol 2020;30:413-24.
86. Abajian A, Murali N, Savic LJ, et al. Predicting treatment response to image-guided therapies using machine learning: an example for trans-arterial treatment of hepatocellular carcinoma. J Vis Exp 2018; doi: 10.3791/58382.
87. Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol 2012;4:a008763.
88. Ziv E, Yarmohammadi H, Boas FE, et al. Gene signature associated with upregulation of the Wnt/β-catenin signaling pathway predicts tumor response to transarterial embolization. J Vasc Interv Radiol 2017;28:349-355.e1.
89. Chen B, Garmire L, Calvisi DF, Chua MS, Kelley RK, Chen X. Harnessing big “omics” data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2020;17:238-51.
90. Wilhelm S, Carter C, Lynch M, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov 2006;5:835-44.
91. Abou-Alfa GK, Schwartz L, Ricci S, et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J Clin Oncol 2006;24:4293-300.
92. Zhu YJ, Zheng B, Wang HY, Chen L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol Sin 2017;38:614-22.
93. Abuhelwa AY, Badaoui S, Yuen HY, et al. A clinical scoring tool validated with machine learning for predicting severe hand-foot syndrome from sorafenib in hepatocellular carcinoma. Cancer Chemother Pharmacol 2022;89:479-85.
94. Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:525-43.
95. Chen D, Liu J, Zang L, et al. Integrated machine learning and bioinformatic analyses constructed a novel stemness-related classifier to predict prognosis and immunotherapy responses for hepatocellular carcinoma patients. Int J Biol Sci 2022;18:360-73.
96. Sangro B, Melero I, Wadhawan S, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol 2020;73:1460-9.
97. Zeng Q, Klein C, Caruso S, et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J Hepatol 2022;77:116-27.
98. Lui TKL, Cheung KS, Leung WK. Machine learning models in the prediction of 1-year mortality in patients with advanced hepatocellular cancer on immunotherapy: a proof-of-concept study. Hepatol Int 2022;16:879-91.
99. Ferrero E, Dunham I, Sanseau P. In silico prediction of novel therapeutic targets using gene-disease association data. J Transl Med 2017;15:182.
100. Tong Z, Zhou Y, Wang J. Identifying potential drug targets in hepatocellular carcinoma based on network analysis and one-class support vector machine. Sci Rep 2019;9:10442.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Panettieri E, Campisi A, Bianchi V, Giuliante F, De Rose AM. Artificial intelligence and its role in guiding liver-directed therapy for hepatocellular carcinoma: Is it ready for prime time?. Hepatoma Res 2022;8:41. http://dx.doi.org/10.20517/2394-5079.2022.57
AMA Style
Panettieri E, Campisi A, Bianchi V, Giuliante F, De Rose AM. Artificial intelligence and its role in guiding liver-directed therapy for hepatocellular carcinoma: Is it ready for prime time?. Hepatoma Research. 2022; 8: 41. http://dx.doi.org/10.20517/2394-5079.2022.57
Chicago/Turabian Style
Panettieri, Elena, Andrea Campisi, Valentina Bianchi, Felice Giuliante, Agostino Maria De Rose. 2022. "Artificial intelligence and its role in guiding liver-directed therapy for hepatocellular carcinoma: Is it ready for prime time?" Hepatoma Research. 8: 41. http://dx.doi.org/10.20517/2394-5079.2022.57
ACS Style
Panettieri, E.; Campisi A.; Bianchi V.; Giuliante F.; De Rose AM. Artificial intelligence and its role in guiding liver-directed therapy for hepatocellular carcinoma: Is it ready for prime time?. Hepatoma. Res. 2022, 8, 41. http://dx.doi.org/10.20517/2394-5079.2022.57
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 34 clicks
Cite This Article 34 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.