Promise and pitfalls of new viral biomarkers for hepatocellular carcinoma risk prediction in patients with chronic hepatitis B
Chronic hepatitis B virus (CHB) infection - estimated to affect 290 million individuals - remains the leading global cause of liver-related mortality and a risk factor for hepatocellular carcinoma (HCC)[1]. Importantly, 56% of liver cancers worldwide were attributable to CHB based on GLOBOCAN estimates from 2012[2]. Current first-line therapies for CHB reduce but do not eliminate the risk of HCC, and for those fortunate enough to experience clearance of hepatitis B surface antigen (HBsAg), either spontaneous or on-treatment, the risk of HCC drops further but again persists[3,4]. Additionally, unlike other etiologies of chronic liver disease, HCC develops in the absence of advanced fibrosis in up to 30% of HBsAg-positive patients[5]. Taken together, these realities dictate that surveillance for HCC is paramount in nearly all circumstances for patients with CHB.
The surveillance pathway adopted by different international hepatology society groups mostly align, with ultrasound +/- serum alpha-fetoprotein every 6 months recommended in all cirrhotics and some CHB at higher risk, such as Asian men over the age of 40, Asian women over the age of 50 and Africans over the age of 20-40[6]. With a global average life expectancy of 73 years, one can imagine that the cumulative burden of surveillance on both individuals and healthcare systems over lifetimes is extraordinarily high. Therefore, a key objective has been to develop risk prediction scores to identify CHB groups at high risk for HCC, but more importantly, those in whom risk is low enough that surveillance may not be indicated. These risk scores typically stratify on treatment status and include demographic factors (e.g., age, sex), laboratory values reflective of liver inflammation or fibrosis (e.g., serum ALT, albumin, platelet count), and - in some instances - viral factors (e.g., HBV DNA, HBeAg status)[6]. Despite the development of numerous validated scores with excellent negative predictive values (> 95%), there has not yet been clinical consensus on their use to exclude patients from HCC surveillance. More recently, quantitative hepatitis B surface antigen levels (qHBsAg), hepatitis B core-related antigen levels (HBcrAg), HBV RNA, and markers of HBV evolution (e.g., preS and basal core mutations) have been examined as adjunct tools for HCC risk prediction - with the hope that addition of these viral biomarkers will improve precision of these scores.
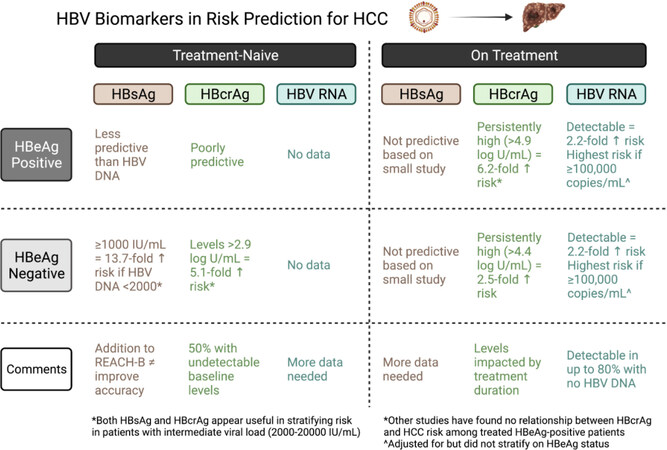
Circulating HBsAg comes from either integrated DNA or cccDNA sources, and quantification of HBsAg is a useful tool in the management of CHB, particularly with respect to predicting the likelihood of HBsAg loss, and is, at present, the most accessible biomarker for commercial use. However, a relationship between qHBsAg and the development of HCC has not been consistently demonstrated. Though qHBsAg correlates with cccDNA in both tumor and non-neoplastic tissue[7], median qHBsAg did not differ between HCC cases and controls in several case-control studies[8-10] nor did qHBsAg predict HCC risk in several observational studies[11,12]. A meta-analysis of eight studies of low heterogeneity, however, determined a pooled 2.5-fold (95%CI: 2.2-2.8) increased risk of HCC with qHBsAg ≥ 1000 IU/mL[13]. Further, neither adding qHBsAg nor replacing HBV DNA with qHBsAg in the original REACH-B score (which predicts HCC risk among non-cirrhotics who are treatment-naïve[14]) resulted in enhanced levels of accuracy[15]. Use of qHBsAg levels among CHB groups with low HBV DNA levels, such as inactive or indeterminate phenotypes, yield better risk prediction. For example, while the receiver operating curve (ROC) for ALT and HBV DNA both outperformed qHBsAg at predicting long-term HCC risk in a large non-cirrhotic CHB cohort, qHBsAg ≥ 1000 IU/mL was strongly associated with HCC among the subgroup of HBeAg-negative with HBV DNA < 2000 IU/mL (HR = 13.7, 4.8-39.3) but notably not for higher viral loads[16]. Among HBeAg-negative patients with an intermediate viral load (HBV DNA 2000-19,999 IU/mL), the area under ROC for 10-year HCC development was 0.68 for a combination of qHBsAg plus HBV DNA vs. only 0.54 for HBV DNA alone[17]. In fact, the group with the lowest annual incidence rate of HCC were those with either a low viral load plus qHBsAg < 1000 IU/mL or intermediate viral load plus qHBsAg < 100 IU/mL at 0.6 cases per 1000 person-years[17]. Thus, qHBsAg has limited application in risk prediction scores for HCC, with the exception of HBeAg-negative CHB with low or intermediate levels of HBV DNA.
HBcrAg has emerged as a potentially more promising viral biomarker for HCC risk prediction. HBcrAg components include HBeAg, hepatitis B core antigen, and a precore protein, with production dependent on the level of transcription/translation of the HBV precore/core gene[18]. Importantly, HBcrAg correlates with intrahepatic cccDNA[19], even among those with low or undetectable serum HBV DNA[20]. This characteristic allows for the use of HBcrAg for risk prediction among treated patients, and both pre-treatment and on-treatment HBcrAg values have been associated with HCC[21-23]. For example, higher HCC risk among HBeAg-negative was observed in those with persistent or on-treatment (at 1 year) HBcrAg >
HBV RNA has recently entered the arena as a viral biomarker for treated CHB patients. HBV RNA remains elevated in up to 78% of CHB patients who achieve undetectable HBV DNA on treatment, much higher than the 30% for HBcrAg[27]. A small case-control study demonstrated that a detectable HBV RNA among treated patients was more common in HCC cases, whereas no difference was seen in qHBsAg levels[10]. In a prospective cohort study of 2974 treated patients (40% HBeAg-positive, 37% with cirrhosis), the 70% of individuals with baseline detectable HBV RNA were 2.2-fold more likely to develop HCC after 4.4 years of follow-up (cumulative incidence of 4.1% at 5 years if detectable compared to 1.8% if not). The greatest risk was observed for those with markedly elevated baseline HBV RNA of ≥ 100,000 copies/mL[28]. The risk of HCC was also 4-fold higher if both HBV RNA and HBV DNA were detectable[28]. Further studies are needed, but initial results are encouraging.
Lastly, HBV variants, specifically those in the preS, basal core promoter (A1762T and G1764A) and precore (G1896A) regions of the HBV genome, have been linked to HCC risk independent of ALT or HBV DNA levels, with the magnitude of risk modulated by HBV genotype and HBeAg status[29-31]. Natural history studies highlight that viral mutations accumulate over an individual’s lifetime of infection; thus, discerning a causative role of specific mutation beyond that related to the inflammatory-fibrotic process caused by chronic viremia can be challenging. In a recent meta-analysis of 21 studies (all among Asian populations), a 2-3-fold elevated HCC risk was demonstrated with the presence of either preS deletion (all, preS1 or preS2) or preS2 start codon mutation[32]. A causative role is suggested by a recent translational study showing that intracellular accumulation of a secretion-defective pre-S2 deletion mutant led to increased stress in the endoplasmic reticulum, calcium overload, mitochondrial dysfunction, and enhanced liver fibrosis[33]. The optimal methods to detect pre-S deletions and pre-S deleted proteins have not been established, and none are available for routine clinical care[34]. Given the heterogeneity (and infrequent testing) of HBV mutations, few have been incorporated into risk prediction scores, but advances in sequencing and machine learning may pave the way for their inclusion in the future.
While more studies are needed, it is improbable that any single viral biomarker will be a panacea for HCC risk prediction for all CHB phenotypes. Rather, the inclusion of viral biomarkers in risk predictor scores will likely be tailored to the patient context - for example, whether cirrhosis is present, HBeAg positive or negative, on treatment or off, and whether the marker itself is detectable (see Figure 1). In fact, we would argue that the use or addition of viral biomarkers will be unlikely to achieve the degree of risk stratification necessary to allow clinicians to identify patients who can forgo surveillance for HCC. Rather, the rapidly evolving field of serum tumor biomarkers, such as methylated DNA or single nucleotide polymorphisms, might better stratify risk, or eventually supplant imaging-based surveillance, thereby simplifying the process and cost of surveillance.
However, these data do suggest a role for viral biomarkers as an adjuvant to decisions regarding the need for antiviral therapy to lower HCC risk. This is particularly relevant for patients in the “grey zone”, for whom specific thresholds of qHBsAg, HBcrAg, or HBV RNA may push one towards initiating antiviral therapy, even if ALT and HBV DNA levels are lower than traditional thresholds for treatment. Further, nearly 80% of indeterminate phase patients have at least one high-risk mutation[35], justifying a potential role in identifying viral variants as part of treatment decision-making. As treatment endpoints shift to HBsAg loss, pre-treatment viral biomarkers might be used to predict HCC risk after seroclearance and potentially influence surveillance strategies. For example, as more patients achieve this favorable outcome at an earlier time point in the duration of their infection (and younger age), the risk of HCC might be so exceedingly low that surveillance after HBsAg loss is not needed. As such, further studies of HBV biomarkers remain paramount, particularly in these special populations and in the face of upcoming new therapies, with the expectation that novel biomarkers will contribute to refinements in HCC prevention and surveillance in the future.
DECLARATIONS
Authors’ contributionsDrafting of the manuscript: Zhou K
Critical revision of the manuscript for important intellectual content: Zhou K, Terrault N
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestZhou K reported institutional grant support from Gilead Sciences. Terrault N disclosed institutional grant support from Gilead Sciences, GSK, and Roche-Genentech and consults for Saol Therapeutics and Moderna.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
1. Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis b virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 2018;3:383-403.
2. Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J Cancer 2018;142:2471-7.
3. Yip TC, Wong GL, Chan HL, et al. HBsAg seroclearance further reduces hepatocellular carcinoma risk after complete viral suppression with nucleos(t)ide analogues. J Hepatol 2019;70:361-70.
4. Kim GA, Lee HC, Kim MJ, et al. Incidence of hepatocellular carcinoma after HBsAg seroclearance in chronic hepatitis B patients: a need for surveillance. J Hepatol 2015;62:1092-9.
5. Desai A, Sandhu S, Lai JP, Sandhu DS. Hepatocellular carcinoma in non-cirrhotic liver: a comprehensive review. World J Hepatol 2019;11:1-18.
6. Wong VW, Janssen HL. Can we use HCC risk scores to individualize surveillance in chronic hepatitis B infection? J Hepatol 2015;63:722-32.
7. Wang Q, Luan W, Warren L, et al. Serum hepatitis B surface antigen correlates with tissue covalently closed circular DNA in patients with hepatitis B-associated hepatocellular carcinoma. J Med Virol 2016;88:244-51.
8. Kawanaka M, Nishino K, Nakamura J, et al. Quantitative levels of hepatitis B virus DNA and surface antigen and the risk of hepatocellular carcinoma in patients with hepatitis B receiving long-term nucleos(t)ide analogue therapy. Liver Cancer 2014;3:41-52.
9. Cheung KS, Seto WK, Wong DK, Lai CL, Yuen MF. Relationship between HBsAg, HBcrAg and hepatocellular carcinoma in patients with undetectable HBV DNA under nucleos(t)ide therapy. J Viral Hepat 2017;24:654-61.
10. Mak LY, Huang Q, Wong DK, et al. Residual HBV DNA and pgRNA viraemia is associated with hepatocellular carcinoma in chronic hepatitis B patients on antiviral therapy. J Gastroenterol 2021;56:479-88.
11. Liang LY, Wong VW, Toyoda H, et al. Serum hepatitis B core-related antigen predicts hepatocellular carcinoma in hepatitis B e antigen-negative patients. J Gastroenterol 2020;55:899-908.
12. Tada T, Kumada T, Toyoda H, et al. HBcrAg predicts hepatocellular carcinoma development: an analysis using time-dependent receiver operating characteristics. J Hepatol 2016;65:48-56.
13. Thi Vo T, Poovorawan K, Charoen P, et al. Association between hepatitis B surface antigen levels and the risk of hepatocellular carcinoma in patients with chronic hepatitis B infection: systematic review and meta-analysis. Asian Pac J Cancer Prev 2019;20:2239-46.
14. Yang H, Yuen M, Chan HL, et al. Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): development and validation of a predictive score. Lancet Oncol 2011;12:568-74.
15. Yang HI, Tseng TC, Liu J, et al. Incorporating serum level of hepatitis B surface antigen or omitting level of hepatitis B virus DNA does not affect calculation of risk for hepatocellular carcinoma in patients without cirrhosis. Clin Gastroenterol Hepatol 2016;14:461-8.e2.
16. Tseng TC, Liu CJ, Yang HC, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology 2012;142:1140-9.e3; quiz e13-4.
17. Tseng TC, Liu CJ, Chen CL, et al. Risk stratification of hepatocellular carcinoma in hepatitis B virus e antigen-negative carriers by combining viral biomarkers. J Infect Dis 2013;208:584-93.
18. Suzuki F, Miyakoshi H, Kobayashi M, Kumada H. Correlation between serum hepatitis B virus core-related antigen and intrahepatic covalently closed circular DNA in chronic hepatitis B patients. J Med Virol 2009;81:27-33.
19. Testoni B, Lebossé F, Scholtes C, et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol 2019;70:615-25.
20. Chen S, Jia J, Gao Y, et al. Clinical evaluation of hepatitis B core-related antigen in chronic hepatitis B and hepatocellular carcinoma patients. Clin Chim Acta 2018;486:237-44.
21. Hosaka T, Suzuki F, Kobayashi M, et al. Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues. Aliment Pharmacol Ther 2019;49:457-71.
22. Kumada T, Toyoda H, Tada T, et al. Effect of nucleos(t)ide analogue therapy on hepatocarcinogenesis in chronic hepatitis B patients: a propensity score analysis. J Hepatol 2013;58:427-33.
23. Honda M, Shirasaki T, Terashima T, et al. Hepatitis B virus (HBV) core-related antigen during nucleos(t)ide analog therapy is related to intra-hepatic HBV replication and development of hepatocellular carcinoma. J Infect Dis 2016;213:1096-106.
24. Mak LY, Ko KL, To WP, et al. Entecavir reduced serum hepatitis B core-related antigen in chronic hepatitis B patients with hepatocellular carcinoma. Gut Liver 2020;14:665-8.
25. Tseng TC, Liu CJ, Hsu CY, et al. High level of hepatitis B core-related antigen associated with increased risk of hepatocellular carcinoma in patients with chronic HBV infection of intermediate viral load. Gastroenterology 2019;157:1518-29.e3.
26. Tseng TC, Hosaka T, Liu CJ, et al. Hepatitis B core-related antigen stratifies the risk of liver cancer in HBeAg-negative patients with indeterminate phase. Am J Gastroenterol 2022; doi: 10.14309/ajg.0000000000001691.
27. Mak LY, Cloherty G, Wong DK, et al. HBV RNA profiles in patients with chronic hepatitis B under different disease phases and antiviral therapy. Hepatology 2021;73:2167-79.
28. Liu S, Deng R, Zhou B, et al. Association of serum hepatitis B virus RNA with hepatocellular carcinoma risk in chronic hepatitis B patients under nucleos(t)ide analogues therapy. J Infect Dis 2021; doi: 10.1093/infdis/jiab597.
29. Liu S, Zhang H, Gu C, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst 2009;101:1066-82.
30. Yang HI, Yeh SH, Chen PJ, et al. REVEAL-HBV Study Group. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 2008;100:1134-43.
31. Wei F, Zheng Q, Li M, Wu M. The association between hepatitis B mutants and hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 2017;96:e6835.
32. Wungu CDK, Ariyanto FC, Prabowo GI, Soetjipto S, Handajani R. Meta-analysis: association between hepatitis B virus preS mutation and hepatocellular carcinoma risk. J Viral Hepat 2021;28:61-71.
33. Liang YJ, Teng W, Chen CL, et al. Clinical implications of HBV PreS/S mutations and the effects of PreS2 deletion on mitochondria, liver fibrosis, and cancer development. Hepatology 2021;74:641-55.
34. Lin YT, Jeng LB, Su IJ, Teng CF. Approaches for detection of hepatitis B virus pre-S gene deletions and pre-S deleted proteins and their application in prediction of higher risk of hepatocellular carcinoma development and recurrence. Viruses 2022;14:428.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Zhou K, Terrault N. Promise and pitfalls of new viral biomarkers for hepatocellular carcinoma risk prediction in patients with chronic hepatitis B. Hepatoma Res 2022;8:15. http://dx.doi.org/10.20517/2394-5079.2022.06
AMA Style
Zhou K, Terrault N. Promise and pitfalls of new viral biomarkers for hepatocellular carcinoma risk prediction in patients with chronic hepatitis B. Hepatoma Research. 2022; 8: 15. http://dx.doi.org/10.20517/2394-5079.2022.06
Chicago/Turabian Style
Zhou, Kali, Norah Terrault. 2022. "Promise and pitfalls of new viral biomarkers for hepatocellular carcinoma risk prediction in patients with chronic hepatitis B" Hepatoma Research. 8: 15. http://dx.doi.org/10.20517/2394-5079.2022.06
ACS Style
Zhou, K.; Terrault N. Promise and pitfalls of new viral biomarkers for hepatocellular carcinoma risk prediction in patients with chronic hepatitis B. Hepatoma. Res. 2022, 8, 15. http://dx.doi.org/10.20517/2394-5079.2022.06
About This Article
Copyright
Data & Comments
Data

 Cite This Article 6 clicks
Cite This Article 6 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.