Involvement of DNA methylation in tissue regeneration upon liver injury
Abstract
Following injury such as partial hepatectomy, the liver activates its response to proliferate and repopulate the lost liver mass. This process is associated with the re-expression of pro-regenerative genes that are controlled epigenetically. While epigenetic control of gene expression can be observed in many forms (DNA methylation, histone modification, chromatin remodeling, etc.), this review will focus on the role of regulators of DNA methylation in the concept of cell proliferation and progression and its relevance to tissue regeneration. Obligate proteins UHRF1 and DNMT1 are key players in the maintenance of methylation in the process of DNA replication. Both proteins have a well-established role in cell proliferation, progression and methylation maintenance, and are indicators of the stress response. Understanding how these regulatory mechanisms function is crucial in determining clinical applications on restoring lost tissue in the liver as a result of infection, toxins, or other liver pathologies.
Keywords
LIVER REGENERATION FOLLOWING INJURY
The liver is an integral organ responsible for various physiological processes. Its role includes macronutrient metabolism, blood volume regulation, immune system support, endocrine control of growth signaling pathways, lipid and cholesterol homeostasis, and the breakdown of xenobiotic compounds, including many current drugs[1]. Understanding how the liver develops, functions, and regenerates are important to reduce the global morbidity and mortality of liver diseases. In 2017, systematic analysis of the global burden of liver disease, deaths due to cirrhosis, and chronic liver disease accounted for 2.4% of total deaths vs 1.9% in 1990[2]. These data suggest a significant increase in the burden of liver disease over the years. Hence, the need to reinforce existing knowledge and treatment paradigm is warranted.
Among mammals, the liver is regarded as a particularly unique organ because it is the only internal organ truly capable of naturally restoring its full mass and functions after injury or resection[3]. In 1931, Higgins and Anderson first demonstrated a liver regenerative ability in a murine model of partial hepatectomy (PH). Following the removal up of around 70% of the liver mass, it completely recovered the hepatic mass in about one week[4]. In 1964, Buccher and Swaffield further demonstrated this phenomenon by incorporating labeled-thymidine into the DNA. The restoration of liver mass and function proved the existence of delicately balanced mechanisms for weighing the numerous metabolic demands against the urgent need for replacement of lost liver tissue[5].
Following PH, the proliferation of quiescent hepatocytes and cholangiocytes, followed by proliferation of hepatic stellate cells and endothelial cells, quickly restores the liver into its original mass[6,7]. This is also the same effective response reaction of the liver tissue upon acute and chronic hepatic damage, caused by toxins or viral infections. This regenerative process is a rapid but tightly controlled proliferation of hepatocytes and other liver cells[8,9]. However, when the hepatic damage is severe, the burden of recovering lost liver mass mobilizes the hepatic progenitor cells (HPC). This so-called “ductular reaction” is activated and is expanded to reverse the injury[10]. These cells rarely acquire age-related genetic defects associated with cancer induction that may imply their protective mechanism against genetic damage[6]. The activation of HPCs was observed in chronic cholestasis, regenerating human liver after sub-massive necrosis, alcoholic liver disease, and focal nodular hyperplasia[11]. Hence, unique hepatic regenerative capacity needs to be explored to understand its mechanistic controls for the application of liver restoration after various types of injuries.
Prox1, a transcription factor containing homeobox and Prospero domain, has been implicated in liver morphogenesis. Prox1 expression is restricted and specific as an early specific marker for the developing liver (and pancreas) in the mammalian foregut endoderm[12], where the loss of Prox1 leads to the formation of a smaller liver[13]. During liver development, however, Prox1 is absent in bile duct epithelial cells and immortalized hepatoblasts, suggesting a conserved function for the specification of embryonic hepatoblasts into mature hepatocytes[14,15]. Following PH and 2-acetyl-aminofluorene treatment, the amount of Prox1 mRNA was found to be reduced, but Prox1 protein remained detectable and rather stable in the nuclei of parenchymal cells, suggesting Prox1 as a stable marker of hepatocytes in normal adult, injured, regenerating rat liver[16].
DNA METHYLATION REGULATION IN THE LIVER
Epigenetics controls the expression of most, if not all, genes. In the concept of liver regeneration, very distinct patterns of gene expression are required to trigger the activation of a quiescent liver to a regenerative one. In the event of liver damage, hepatocytes turn on a pro-regenerative switch to undergo synchronized cell proliferation and restore liver mass[3]. Since epigenetics takes part in the maintenance and control of gene expression, its function in liver regeneration is also pivotal.
Epigenetics is a complex combination of DNA modifications, histone variants, histone post-translational modifications, non-coding RNAs, chromatin remodelers, and other factors[17]. This regulatory mechanism takes over the control of turning on genes programmed to promote regeneration. The precise programming controls the transition of a quiescent hepatocyte to a proliferative hepatocyte[3]. All the epigenetic processes remodel the chromatin, thereby allowing access of transcription factors on the repressed genes in the quiescent liver to be activated and to signal cell cycle progression[18].
During liver development in the mouse model, most epigenetic modifiers, such as enzymes involved in DNA methylation, histone modifications, and chromosome-remodeling factors, are highly expressed in the perinatal stage and will regress by postnatal day 21. This downregulation is accompanied by a decrease in hepatocyte proliferation, the achievement of a fully differentiated state, and the establishment of proper liver architecture[19]. The cells in the adult liver therefore are efficiently coordinated by transcription factors and chromatin regulators of terminal differentiation[20]. It is important to note that the cells in a regenerative and developing liver maintain a delicate balance between the need to proliferate and the maintenance of a differentiated function. Therefore, these two physiological liver states share an overlap in the genes involved in cell growth and differentiation[21,22]. Hence decoding the epigenetic information of hepatocytes in a developing liver could reflect what is happening in a regenerating one.
DNA methylation is one of the most characterized forms of epigenetic modifications. It controls gene expression through the alteration of chromosomal structure, DNA conformation and stability, and the interaction between DNA and protein[23]. It involves the transfer of a covalent methyl group to the C5 position of cytosine to form 5-methylcytosine by DNA methyltransferases (DNMTs). During organogenesis, its signature changes due to a dynamic process involving both de novo methylation and demethylation. Therefore, differentiated cells develop a stable and unique pattern that regulates tissue-specific gene transcription[24].
Obligate partners of DNA methylation, ubiquitin-like plant homeodomain, and RING finger domain 1 [UHRF1/NP95/ICBP90 (inverted CCAAT box-binding protein 90)] and DNA methyltransferase 1 (DNMT1) were found to be key players associated with the methylation maintenance of genes and cell proliferation in an adult hepatocyte. In the event of hepatic injury, these players were crucial in ensuring the methylation integrity of genes in the process of replication[25]. While there is limited information on how this cascade happens, this review will briefly describe the significance of the DNA methylation regulators DNMT1 and UHRF1 in liver regeneration following liver injury.
DNMTs
DNMT domains are structurally conserved and make up a highly regulated family, majorly composed of the canonical enzymes DNMT1, DNMT3A, and DNMT3B, while DNMT2 consists of only a catalytic domain. DNMT2 was shown to lack DNA methyltransferase activity and functions instead as a transfer RNA methyltransferase[26]. The structure and function of the DNMT family were previously reviewed in[27-29].
Due to the key roles of DNMTs in cell cycle and progression, their participation in embryo development and tissue regeneration is important to understand. Their tight regulation is very specific in the cell cycle stages. For instance, DNMT3a was consistently downregulated throughout cell cycle phases, while DNMT1 and DNMT3b were induced in the S-phase stage[30]. Considering their roles in cell cycle progression, their differentially regulated expression was expected to be indicative of cell proliferation and progression. During embryo development and tissue regeneration, both DNMT1 and DNMT3b were expected to be upregulated[18]. Accordingly, DNMT1 was shown to be upregulated by hepatocyte growth factor, a key mitogenic player in liver regeneration[18].
One of the main roles of DNMT1 is the maintenance of DNA methylation patterns in the daughter cells, which it can perform via the binding and the recognition of UHRF1 hemimethylated DNA in the replication fork[31]. DNMT1 contains a carboxyterminal domain that serves as the catalytic function of the enzyme and an N-terminal regulatory domain which has an independent transcription repressor activity[32]. Moreover, DNMT1 also has the capacity for de novo methylation[33]. The role of DNMT1 was not found to be crucially important in mature hepatocytes but was critical during physiological processes involving liver development and regeneration[18].
DNMT1 is an essential regulator maintaining both epigenetic reprogramming during DNA replication and genome stability. While it is commonly associated with full methylation of a hemimethylated substrate, there have been several reports of de novo methylation activities for DNMT1[34]. This is attributed to the epigenetic ability of DNMT1 to cooperatively establish tissue-specific methylation patterns. Kaji et al.[35] were able to generate Dnmt1 knockout mice (Dnmt1Δalb). They showed that Dnmt1 loss in postnatal hepatocytes (4‐week-, 8‐week-, and 20‐week‐old mice) was largely associated with gene de-repression, including reactivation of imprinted genes associated with cell cycle regulation, immune response, DNA repair, cytoskeletal arrangement, and cell adhesion. Effective knockdown of Dnmt1 in cultured HPCs caused severe DNA damage, cell cycle arrest, senescence, and cell death.
DNMT1 loss is also a helpful indicator of replication stress, preventing further expansion of epigenetically damaged cells. Unterberger et al.[36] showed that DNMT1 depletion activated a cascade of genotoxic stress checkpoint proteins, resulting in phosphorylation of checkpoint kinases. Milutinovic et al.[37] further suggested that an intra-S-phase arrest was triggered by the reduction of DNMT1 and not by demethylation that happens after DNMT1 knockdown. The silencing of DNMT1 induced a set of genes associated with genotoxic stress response such as NF-κB, JunB, ATF-3, and GADD45β (growth arrest DNA damage 45β gene). Their results were able to provide information on the distinct role of DNMT1 also as a stress indicator of epigenetically damaged DNA preventing it from expansion to properly coordinate genetic and epigenetic inheritance in DNA replication.
UHRF1
UHRF1 is a multi-domain nuclear regulator of epigenetic modification in the recognition of histone marks through subsequent interactions with chromatin-modifying proteins and in the maintenance of DNA methylation[38]. The functions of UHRF1 and DNMTs are associated with each other, where UHRF1 is required for both DNMT1 localization and stability[39]. The functional activity, specificity and degradation of DNMT1 are mainly promoted by UHRF1[40]. It is also important to note that UHRF1 is identified as a regulator of cell proliferation in several tissues and is an equally important regulator of hepatocyte proliferation[41]. While there is still little information about the role of UHRF1 in liver cell proliferation and apoptosis, its role as an epigenetic regulator is well characterized[42].
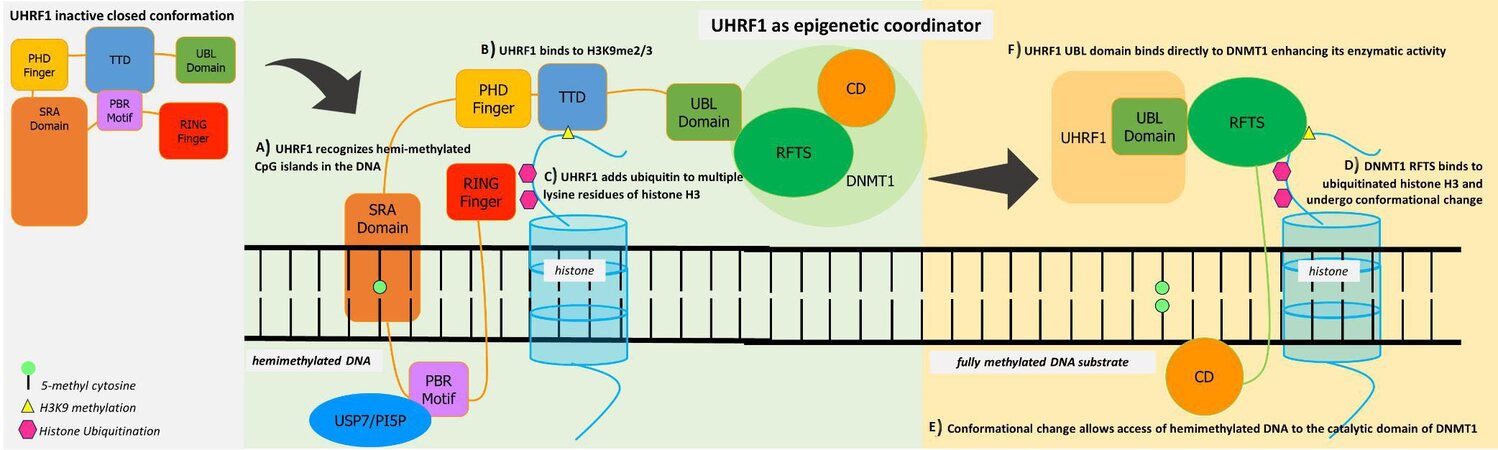
As an epigenetic coordinator, the different domains of UHRF1 are involved in methylation and histone alteration. The following domains compose its structure: ubiquitin-like (UBL) domain, tandem Tudor domain (TTD), and plant homeodomain (PHD), polybasic region (PBR), SET and RING associated (SRA) domain, and the really interesting new gene RING finger. The closed inactive form of UHRF1 can be activated by the binding and phosphorylation of phosphatidylinositol 5-phosphate (PI5P) which releases PBR from its binding to TTD. The conformational switch opens the protein complex and results in the recognition and binding of SRA to the hemimethylated DNA. This binding allows the effective identification of regions of DNA that need to be fully methylated[31,43,44].
The conformational change also allows the interaction of the TTD-PHD complex with methylated H3K9 and ubiquitination of the histone H3 through the UHRF1 RING finger. These series of processes allow the effective recruitment of DNMT1 to DNA replication sites. The RFTS domain of DNMT1 interacts with the UBL domain of UHRF1, to release it from its self-inhibitory activity. It binds to the ubiquitinated histone H3 and undergoes a conformational change as well. This in turn gives access to the catalytic domain of DNMT1. Access to the catalytic domain is necessary for methylation in the newly replicated DNA, resulting in a fully methylated DNA substrate. This process takes effect during the S-phase of the cell cycle. Upon completion of DNA replication and methylation inheritance, the cell cycle proceeds to the G2 phase where both these macromolecules undergo degradation. Acetylation and ubiquitination of DNMT1 and UHRF1 lead to their decrease, ensuring the cell’s normal cycle[43,44] [Figure 1].
Figure 1. UHRF1 acts as a multifunctional epigenetic regulator allowing crosstalk between DNA methylation and histone code. Its closed conformation from the binding of TTD and PBR can be activated by the binding of USP7 or PI5P (phosphatidylinositol 5-phosphate) to the PBR motif. (A) UHRF1 SRA domain identifies and recognizes hemimethylated sites of DNA. (B) PHD finger and TTD complex recognize methylated H3K9, which allows proper localization of DNMT1 on genomic loci. (C) RING finger function such as of ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys23 and Lys18. (D) These ubiquitin markers are recognized by the DNMT1 RFS, promoting its localization to replication foci, which disrupts the interaction of RFTS and the catalytic domain of DNMT1. (E) This conformational change allows access of hemimethylated DNA to the catalytic domain of DNMT1, which results in a fully methylated DNA substrate. (F) The UBL domain of UHRF1 binds directly to DNMT1, further enhancing its enzymatic activity towards newly replicated chromatin. The mechanistic coordinated roles of both proteins ensure the faithful propagation of the DNA methylation patterns in DNA replication. Illustration was based on[43,44].
Most UHRF1 knockout studies in mammals resulted in early gestation death, suggesting the role of UHRF1 in cell growth and proliferation in tissues and organs. In 2004, Obata et al.[45] introduced UHRF1 tissue-specific knockout studies. By using knockout in vitro in breast cancer cells, the loss of UHRF1 resulted in different phenotypes including cell cycle arrest[46], cell cycle arrest in G2/M-phase and apoptosis dependent on caspase 8[47], loss of contact inhibition[48], and sensitivity to cancer treatment through its interaction with TopoIIα[49].
In 2007, the group of Sadler identified the UHRF1 gene (uhrf1 in zebrafish) essential in hepatic growth and embryonic survival[41]. Taking advantage of “true” tissue regeneration by PH indicated the zebrafish as a good model for liver regeneration study. About 67% of overall amino acid identity is shared between zebrafish and human UHRF1 and a stronger homology between the structural domains, making the function of the protein in the two organisms significantly conserved. The homology between their structural domains has also been associated with important epigenetic processes[42].
In this study, the hepatic bud of the uhrf1 mutant zebrafish formed but did not expand and resulted in a small, unilobular, ball-shaped liver, with the animals dying during the larval period. In this model, uhrf1 expression was drastically upregulated during regeneration, in parallel with ccna2 expression, also noticed in mouse liver. Following PH, 100% of the control zebrafishes had substantial regeneration, whereas only 22% of the uhrf1 mutants had any re-growth 5 days after PH. Their data suggest that uhrf1 and ccna2 are required in hepatocyte proliferation in vivo. The researchers postulated that this function was liver-specific since they did not find any associations of uhrf1 with fin regeneration and wound healing[41].
Another study from Jacob’s group[50] showed that the uhrf1 mutants had significantly upregulated genes regulating DNA replication and S phase, despite their reduced liver size. They were also able to observe many cells undergoing DNA replication, indicating that the mutant cells undergo re-replication or an unscheduled DNA replication eventually leading to cell cycle arrest and apoptosis. Global hypomethylation was also observed in the uhrf1 mutant zebrafish which preceded the replication phenotype leading to hepatic outgrowth failure. These in turn can induce DNA damage or genomic instability[51]. Hence, the loss of uhrf1 led to the activation of a cell cycle checkpoint which can be induced by either re-replication or by global hypomethylation. These important data indicated how a cascade of epigenetic disruption affects overall DNA methylation, replication, and apoptosis. The resulting DNA damage checkpoint is an effective mechanism restricting the propagation of epigenetically damaged cells.
A transcriptomic and epigenomic profiling of genes that are differentially expressed in the liver at varied time points after PH in mouse was reported recently[25]. A particular set of genes were observed to peak between 40 and 48 h after PH and had the lowest expression in quiescent livers compared to other differentially expressed genes. Included in this set are UHRF1 and DNMT1, previously reported to have epigenetic functions[41,50]. Further investigation confirmed a significant increase in the expression of the two markers 40 h after PH, and then returning to baseline level[25]. This pattern strongly indicates the active roles of UHRF1 and DNMT1 in liver regeneration.
However, the deletion of UHRF1 in hepatocytes (Uhrf1HepKO), despite indicating global hypomethylation, did not show hypomethylation effects on gene expression, transposon repression, or liver homeostasis. This compensation was attributed to the redistribution of H3K27me3 from promoters to transposons, leading to silencing and further alleviating repression of liver regeneration genes[25]. This information thus provides additional information on the control of gene expression via epigenetic regulation.
INTERACTION BETWEEN DNMT1 AND UHRF1
Several studies suggest a further association of UHRF1 and DNMT1, since they display similar phenotypes in knockout models[36,37,52-54]. In the adult liver, the same downregulated expression of DNMT1 was observed in UHRF1[55]. The dysregulation in these two epigenetic players can lead to disruption of their homeostatic relationship. In the work of Kent et al.[56], both overexpression of UHRTF1 (WT-UHRF1) and an increase in non-phosphorylatable UHRF1 (UHRF1S661A) caused DNA hypomethylation, but acting together with DNMT1, there was induction of an asymmetric epiboly phenotype, ultimately disrupting gastrulation. DNMT1 knockdown suppressed the phenotype caused by UHRF1S661A overexpression, and UHRF1 knockdown suppressed the effect of DNMT1 overexpression. Hence, the stability of DNMT1 is dependent on the phosphorylation of UHRF1, and the crosstalk between the proteins is important at least in zebrafish gastrulation. Such evidence strengthens the relationship between the two proteins as epigenetic regulators.
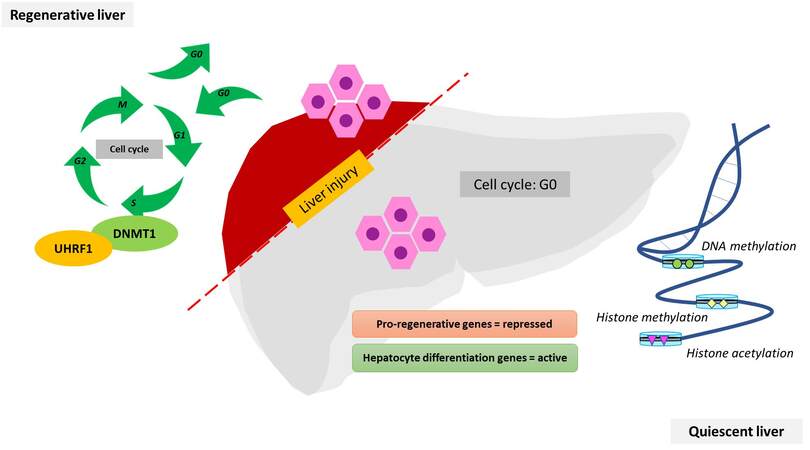
In liver regeneration, this methylation maintenance role of UHRF1 and DNMT1 is very pivotal. In a normal quiescent liver, hepatocytes do not undergo a cell cycle. Pro-regenerative genes are repressed while differentiation genes are active. Upon liver injury, a proliferation switch is turned on in hepatocytes to compensate for the loss of tissue. This dictates the cells to enter the cell cycle phase. During the cell cycle, DNA replication happens and the maintenance mechanism of both UHRF1 and DNMT1 is necessary to ensure the proper methylation pattern throughout the regenerative process [Figure 2].
Figure 2. Involvement of UHRF1 and DNMT1 in a regenerative liver after hepatic injury. The hepatocytes of the quiescent liver do not undergo cell division. Epigenomic modulators such as DNA methylation and histone acetylation and methylation maintain the integrity of repressed and active genes. Upon liver injury, the hepatocytes synchronously re-enter the cell cycle and proliferate, where UHRF1 and DNMT1 work in coordination to maintain the methylation integrity. These regulators display the same temporal patterns as genes that control hepatocyte proliferation. Both are crucial in the DNA methylation machinery.
Recent data also highlighted the correlation of DNMT1 and UHRF1 with euchromatic histone‐lysine methyltransferase 2 (EHMT2/HMT G9a), a gene encoding a methyltransferase that methylates lysine residues of histone H3. In advanced liver injury such as hepatocellular carcinoma (HCC), a significant and correlative overexpression of G9a, DNMT1, and UHRF1 in HCCs is associated with poor prognosis in the patients[57]. Dual epigenetic targeting of DNMT1 and G9a was demonstrated as a promising strategy not only for HCC treatment[57], but also for its earlier event of liver fibrosis[58]. In a mouse model, it was shown that G9a mediated H3K9 dimethylation of target promoters of Ccl5 and Cxcl10, which were aggravated in liver fibrosis[59].
Considering their key roles in development and cell proliferation and progression, we could assume that these molecules are upregulated in the proliferative phase of this physiological process[18]. The need to proliferate while maintaining a differentiated function is equally important in both the developing and regenerating liver[8]. These two physiological states share key regulating genes that function in cell growth and differentiation[21,22]. Hence understanding the epigenetic contribution of hepatocyte proliferation during liver development can shed light on understanding liver regeneration as well.
To date, only limited information is available on the complete signatures of DNA methylation after liver injury due to PH. However, it is important to note that the Ras/Raf/MEK/ERK, JAK/STAT3, and PI3K/Akt pathways are all involved in liver regeneration[60]. Key molecules of these pathways are associated with cell proliferation, growth, and migration. Therefore, the activation and re-expression of these key players might be controlled by DNA methylation or other epigenetic mechanisms.
FUTURE DIRECTION
The regulators of DNA methylation UHRF1 and DNMT1 ensure how a transitioned liver cell will undergo efficient replication and maintain methylation patterns. The close association of UHRF1 and DNMT1 is important in understanding not only cell proliferation according to its implications for a developing organ and a regenerative tissue. Dysregulated UHRF1 and DNMT1 have been associated with developmental regression, fatality, and other liver pathologies, including cancer.
While there is growing knowledge on how UHRF1 regulates the activity, specificity, and degradation of DNMT1, the exact mechanism on how the two regulators can dictate cell cycle arrest, apoptosis, and stress response still needs to be understood. There is a need to fill existing gaps in the understanding of how these molecular players can trigger a fail-safe response in the event of dysregulation. Also, the role of DNMT1 in the maintenance of DNA methylation has been identified to be a crucial part of regeneration, and its role in the turning on and off of genes in the context of a regenerative cell has not been adequately elucidated. Another gap that needs to be fulfilled.
In studying the complexity of regeneration, the zebrafish as a model shows evidence of shared homology, making it a good model to understand liver regeneration. However, the utilization of advanced tools to be able to study this physiological process in a more advanced organism might provide additional information that could apply to humans.
Since the roles of both UHRF1 and DNMT1 have been identified in terms of cell proliferation and maintenance of methylation and as markers of the stress response, this could be meaningful information that can help in the utilization of these mechanisms in the clinical setting. The need to understand molecular concepts underlying tissue regeneration can be proven helpful in clinical applications through (1) improving the category of the patients selected for hepatectomy or liver transplantation; (2) identification of a molecular target on a specific pathway; and (3) developing a so-called artificial liver for therapy in tissue bioengineering.
CONCLUSION
DNA methylation is an epigenetic mechanism takes part in the regulation of gene expression. In the concept of liver regeneration, the coordinated switch on the expression of genes allows the transition of a quiescent liver to a pro-regenerative form. This unique capacity of the liver to heal itself after loss of tissue is an important physiological process governed by cell transformation, proliferation and progression. While DNA methylation is commonly associated with the turning on and off of genes and repression of transposable elements, this review focused on the regulators of DNA methylation: UHRF1 and DNMT1. These two molecular players ensure how a transitioned liver cell will undergo replication and maintain its methylation pattern. The close relationship and shared characteristics of UHRF1 and DNMT1 are important in understanding not only cell proliferation but also their implications for a regenerative cell.
DECLARATIONS
Authors’ contributionsConceived the idea: Sukowati CHS.
Wrote the manuscript: Cabral LKD, Tiribelli C, Sukowati CHS
Read and approved the manuscript: Cabral LKD, Tiribelli C, Sukowati CHS
Availability of data and materialsNot applicable.
Financial support and sponsorshipCHCS was awarded a 2020 grant from the Fondazione Umberto Veronesi, Milan, Italy, and LKDC a fellowship from the Department of Science and Technology and the Philippine Council for Health Research and Development (DoST-PCHRD). The work was partly supported by a grant from Regione Autonoma FVG-Progetti internazionali 2020 (DGR 2195 dd 20/12/2019) to the FIF.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
2. Sepanlou SG, Safiri S, Bisignano C, et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2020;5:245-66.
3. Thakur A, Hoodless PA. Repressive Epigenetic Signatures Safeguard the Liver. Dev Cell 2019;50:3-4.
4. Higgins G. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol 1931;12:186-202.
5. Bucher NLR, Swaffield MN. The rate of incorporation of labeled thymidine into the deoxyribonucleic acid of regenerating rat liver in relation to the amount of liver excised. Cancer Res 1964;24:1611-25.
6. Mishra L, Banker T, Murray J, et al. Liver stem cells and hepatocellular carcinoma. Hepatology 2009;49:318-29.
9. Michalopoulos GK. Hepatostat: Liver regeneration and normal liver tissue maintenance. Hepatology 2017;65:1384-92.
10. Roskams TA, Theise ND, Balabaud C, et al. Nomenclature of the finer branches of the biliary tree: canals, ductules, and ductular reactions in human livers. Hepatology 2004;39:1739-45.
11. Sell S. Comparison of liver progenitor cells in human atypical ductular reactions with those seen in experimental models of liver injury. Hepatology 1998;27:317-31.
12. Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev 2002;118:147-55.
13. Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet 2000;25:254-5.
14. Dudas J, Papoutsi M, Hecht M, et al. The homeobox transcription factor Prox1 is highly conserved in embryonic hepatoblasts and in adult and transformed hepatocytes, but is absent from bile duct epithelium. Anat Embryol (Berl) 2004;208:359-66.
15. Papoutsi M, Dudas J, Becker J, et al. Gene regulation by homeobox transcription factor Prox1 in murine hepatoblasts. Cell Tissue Res 2007;330:209-20.
16. Dudas J, Elmaouhoub A, Mansuroglu T, et al. Prospero-related homeobox 1 (Prox1) is a stable hepatocyte marker during liver development, injury and regeneration, and is absent from "oval cells". Histochem Cell Biol 2006;126:549-62.
17. Macchi F, Sadler KC. Unraveling the Epigenetic Basis of Liver Development, Regeneration and Disease. Trends Genet 2020;36:587-97.
18. Arechederra M, Berasain C, Avila MA, Fernández-Barrena MG. Chromatin dynamics during liver regeneration. Semin Cell Dev Biol 2020;97:38-46.
19. Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD. Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing. Epigenetics 2012;7:914-29.
20. Berasain C, Avila MA. Regulation of hepatocyte identity and quiescence. Cell Mol Life Sci 2015;72:3831-51.
21. Kelley-Loughnane N, Sabla GE, Ley-Ebert C, Aronow BJ, Bezerra JA. Independent and overlapping transcriptional activation during liver development and regeneration in mice. Hepatology 2002;35:525-34.
22. Jochheim-Richter A, Rüdrich U, Koczan D, et al. Gene expression analysis identifies novel genes participating in early murine liver development and adult liver regeneration. Differentiation 2006;74:167-73.
23. Wang S, Wu W. . DNA Methylation Alterations in Human Cancers. Epigenetics in Human Disease 2nd ed. 2018; p. 109-139.
24. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23-38.
25. Wang S, Zhang C, Hasson D, et al. Epigenetic Compensation Promotes Liver Regeneration. Dev Cell 2019;50:43-56.e6.
26. Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006;311:395-8.
27. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 2018;19:81-92.
28. Gowher H, Jeltsch A. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem Soc Trans 2018;46:1191-202.
29. Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure 2008;16:341-50.
30. Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res 2000;28:2108-13.
31. Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007;317:1760-4.
32. Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development 2008;135:1295-302.
33. Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998;19:219-20.
34. Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 2012;335:709-12.
35. Kaji K, Factor VM, Andersen JB, et al. DNMT1 is a required genomic regulator for murine liver histogenesis and regeneration. Hepatology 2016;64:582-98.
36. Unterberger A, Andrews SD, Weaver IC, Szyf M. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol Cell Biol 2006;26:7575-86.
37. Milutinovic S, Zhuang Q, Niveleau A, Szyf M. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J Biol Chem 2003;278:14985-95.
38. Kim KY, Tanaka Y, Su J, et al. Uhrf1 regulates active transcriptional marks at bivalent domains in pluripotent stem cells through Setd1a. Nat Commun 2018;9:2583.
39. Li T, Wang L, Du Y, et al. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res 2018;46:3218-31.
40. Bashtrykov P, Jankevicius G, Jurkowska RZ, Ragozin S, Jeltsch A. The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J Biol Chem 2014;289:4106-15.
41. Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A 2007;104:1570-5.
42. Chu J, Loughlin EA, Gaur NA, et al. UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is required for zebrafish embryogenesis. Mol Biol Cell 2012;23:59-70.
43. Xue B, Zhao J, Feng P, Xing J, Wu H, Li Y. Epigenetic mechanism and target therapy of UHRF1 protein complex in malignancies. Onco Targets Ther 2019;12:549-59.
44. Xie S, Qian C. The Growing Complexity of UHRF1-Mediated Maintenance DNA Methylation. Genes (Basel) 2018;9:600.
45. Obata Y, Furusawa Y, Endo TA, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol 2014;15:571-9.
46. Li X, Meng Q, Rosen EM, Fan S. UHRF1 confers radioresistance to human breast cancer cells. Int J Radiat Biol 2011;87:263-73.
47. Tien AL, Senbanerjee S, Kulkarni A, et al. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J 2011;435:175-85.
48. Hopfner R, Mousli M, Oudet P, Bronner C. Overexpression of ICBP90, a novel CCAAT-binding protein, overcomes cell contact inhibition by forcing Topoisomerase II Alpha expression. Anticancer Res 2002;22:3165-70.
49. Arima Y, Hirota T, Bronner C, et al. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells 2004;9:131-42.
50. Jacob V, Chernyavskaya Y, Chen X, et al. DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos. Development 2015;142:510-21.
51. Truong LN, Wu X. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol 2011;3:13-22.
52. Chen T, Hevi S, Gay F, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet 2007;39:391-6.
53. Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res 2005;65:8635-9.
54. Vijayaraghavalu S, Dermawan JK, Cheriyath V, Labhasetwar V. Highly synergistic effect of sequential treatment with epigenetic and anticancer drugs to overcome drug resistance in breast cancer cells is mediated via activation of p21 gene expression leading to G2/M cycle arrest. Mol Pharm 2013;10:337-52.
55. Fujimori A, Matsuda Y, Takemoto Y, et al. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm Genome 1998;9:1032-5.
56. Kent B, Magnani E, Walsh MJ, Sadler KC. UHRF1 regulation of Dnmt1 is required for pre-gastrula zebrafish development. Dev Biol 2016;412:99-113.
57. Bárcena-Varela M, Caruso S, Llerena S, et al. Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma. Hepatology 2019;69:587-603.
58. Barcena-Varela M, Paish H, Alvarez L, et al. Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. Gut 2021;70:388-400.
59. Wang Z, Cao D, Li C, Min L, Wang G. Mediator MED23 regulates inflammatory responses and liver fibrosis. PLoS Biol 2019;17:e3000563.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Cabral LKD, Tiribelli C, Sukowati CHC. Involvement of DNA methylation in tissue regeneration upon liver injury. Hepatoma Res 2021;7:41. http://dx.doi.org/10.20517/2394-5079.2020.127
AMA Style
Cabral LKD, Tiribelli C, Sukowati CHC. Involvement of DNA methylation in tissue regeneration upon liver injury. Hepatoma Research. 2021; 7: 41. http://dx.doi.org/10.20517/2394-5079.2020.127
Chicago/Turabian Style
Cabral, Loraine Kay D, Claudio Tiribelli, Caecilia H C Sukowati. 2021. "Involvement of DNA methylation in tissue regeneration upon liver injury" Hepatoma Research. 7: 41. http://dx.doi.org/10.20517/2394-5079.2020.127
ACS Style
Cabral, LKD.; Tiribelli C.; Sukowati CHC. Involvement of DNA methylation in tissue regeneration upon liver injury. Hepatoma. Res. 2021, 7, 41. http://dx.doi.org/10.20517/2394-5079.2020.127
About This Article
Copyright
Data & Comments
Data

 Cite This Article 9 clicks
Cite This Article 9 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.