Liver regeneration: metabolic and epigenetic regulation
Abstract
The liver is bestowed with an extraordinary regenerative capability, which is accomplished by a well-coordinated cellular and molecular response at different phases of regeneration. Metabolism, as the primary function of liver, displays various alterations as a consequence of hepatic insufficiency from an injury. These metabolic perturbations are physiologically relevant for promoting hepatocellular proliferation and regeneration. On the other hand, proliferation of otherwise quiescent hepatocytes and accompanied regeneration are regulated by transient, but precisely regulated transcriptional reprogramming. This phase- cell- and time-specific gene expression is controlled by epigenetic mechanisms. Hence, both metabolic and epigenetic changes regulate liver regeneration events. But the cross-talk between metabolic and epigenetic changes for a successful liver regeneration needs to be explored. Since most of the enzymatic players of epigenetic mechanisms rely upon metabolites for their substrates and co-factors, we expect a highly coordinated inter-dependence between metabolism and epigenetics during liver regeneration too. In the present review, we discuss various metabolic and epigenetic regulatory mechanisms for liver regeneration, and put forward the possible metabolic-epigenetic-liver regeneration link for a better understanding of the process and identification of novel targets for liver-related diseases in clinical settings.
Keywords
Introduction
The vital functions of digestion, absorption, detoxification and synthesis of plasma proteins make the liver an indispensable organ of our body. While performing these essential functions, the liver gets exposed to toxin-rich blood from the gut, and thus, to the associated damage to its cells. This might be the reason why liver possesses a unique and extraordinary regenerative power. Though liver regeneration is an extensively studied phenomenon for decades because of its clinical importance, yet, in liver associated diseases that are the leading cause of mortality worldwide[1,2], regeneration failure remains the primary cause. Success of liver transplants, the only cure for end stage liver treatment, relies on the liver’s regenerative ability of both the living-donor and the recipient. Similarly, patients undergoing surgical resection for the treatment of hepatocellular carcinoma[3] also depend on the regenerative success of the remnant liver. In the light of these clinical examples, it is imperative to understand the mechanism underlying successful liver regeneration. A liver regeneration event can be mediated by either hepatocytes or progenitor cells, depending on the severity of injury[4]. The sequence of events is broadly categorized into the phases of priming, proliferation, and termination[5,6]. Each phase is executed by a synchronous and well-coordinated interplay of several cellular and molecular players[7,8]. Over the past few decades, a large number of studies have revealed these players and their roles in the kinetics of liver regeneration. However, the inter-connection/dependence of these players need to be elaborated in order to understand the regeneration process better and develop specific therapeutics. One such provocative, yet unproven link, is between metabolism and epigenetics during liver regeneration[9]. Metabolic perturbations during liver regeneration and phase-specific gene expression via epigenetic mechanisms are two important branches. Recently, many studies have shown that metabolic alterations and epigenetic mechanisms, particularly the histone modifications and DNA methylation are inter-connected during liver regeneration. In the present review, we have summarized recent novel insights in the metabolic and epigenetic regulation of liver regeneration and their interplay thereof.
Liver regeneration: models and mechanisms
The liver is composed of various specialized cell types, of which hepatocytes are the major ones. Hepatocytes remain in proliferative quiescence, but can start dividing if stimulated. Upon injury, hepatocytes can enter the cell cycle and restore the structural and functional hepatic loss by compensatory hyperplasia. But, if the injury is severe, liver specific stem/progenitor cells play the major role in regeneration. Hence, depending on the severity of damage and cell types involved, liver regeneration is of two types, hepatocyte mediated or progenitor/stem cell mediated[4]. The progenitor cell mediated liver regeneration is executed when regeneration by mature hepatocytes is impaired. Compared to stem/progenitor cell mediated regeneration, hepatocyte mediated compensatory hyperplasia is well characterized[4]. In order to dissect the underlying mechanism of liver regeneration, several model systems have been developed over the past few decades.
Experimental paradigms to study liver regeneration
A wide variety of model systems are available to study different aspects of liver regeneration. While in vitro studies using hepatocyte cell cultures are used to investigate liver regeneration specific signalling pathways, in vivo studies are used to get insights into the complex interactions between various hepatic cell types. Recently, long-term three dimensional organoid culture systems for hepatocytes and cholangiocytes from mice and humans have also been established, which recapitulate morphological, functional and transcriptional features of liver regeneration[10]. These organoid systems have opened up experimental avenues for regenerative medicine, disease modeling, gene therapy, and toxicology studies related to the liver[11]. In animal model systems, the first step to provoke regeneration response in otherwise rarely proliferating healthy liver is to induce an injury. Thus, depending on the type of injury, two broadly classified animal models of liver regeneration are: surgical resection and toxin-induced injury[12]. Both of them can be used in animals with a wide range of size, from the zebra fish to pigs[12,13]. The small-sized animals provide financial, logistical, and ethical advantages, but are less suitable for clinical studies due to their differences in size, anatomy and liver metabolism from humans. On the other hand, the larger animals are anatomically and physiologically more similar to humans and provide clinically relevant results, but suffer from logistical, financial, and ethical disadvantages. Thus, both small and large sized animals have their own advantages and disadvantages, and thus, the ultimate choice depends upon the research question being addressed.
Liver regeneration models can also be grouped according to the cell type that is induced to proliferate: liver progenitor cell independent and liver progenitor cell dependent[14]. The liver progenitor cells - hepatic oval cells in rodents and intermediate hepatobiliary cells in humans - mediate liver regeneration only in cases of severe injuries and hepatocyte replication failure[15]. Otherwise, self-replication of hepatocytes contributes to liver regeneration with very little or no contribution of progenitor cells[16].
The commonly used surgical methods of liver regeneration are partial hepatectomy and portal ligation. In partial hepatectomy, upon surgical resection of two-thirds of the liver, the volume of the remaining liver segments increases and gets restored within seven days. This regeneration is more functional than the complete anatomical restoration[4]. The method of two-third partial hepatectomy in rats was first devised by Higgins et al.[17] in 1931; but because the surgery is easy to perform, is reproducible, and well tolerated[6], it has been employed in mice[18], dogs[19], pigs[20,21] and monkeys[22]. Partial devascularization that involves ligating portal vein branches to specific lobes, has been found to cause atrophy of the ligated or portal-deprived lobes, and concomitant compensatory growth of non-occluded residual liver[23]. The portal branch ligation model is simple and reproducible, if adequate operative care is given. In contrast to partial hepatectomy, it is reversible[12] and has been used in mice[24], dogs[25], pigs[26] and monkeys[27]. Though surgical methods are widely used for inducing liver regeneration, their outcome is influenced by a number of factors, including the age of animals[28], fasting before surgery[29], time of surgery[30], and anaesthetics used[29]. Hence, all these experimental variables must be duly considered while using surgery-based liver regeneration models.
The toxin-based, pharmacogical or hepatotoxic models, are relatively easy to perform and are clinically more relevant. However, these models also have various drawbacks such as the lack of reproducibility; variation in regenerative response with respect to dose and mode of administration of the drug; species; age; and the nutritional status of animals[12,31]. Some of these toxin-induced models are also the models for intrinsic drug-induced liver injury (DILI) studies; in fact, acetaminophen (APAP) overdose is one of the most common models for DILI[32]. Some of the commonly used hepatotoxins for liver injury and subsequent regeneration are summarized in Table 1.
Commonly used drugs for hepatotoxic/pharmacological liver regeneration models
| S. No. | Drug/ Hepatotoxin | Principal Cell Affected | Effect |
|---|---|---|---|
| 01 | Carbon tetrachloride | Hepatocytes | Cell membrane damage, dependent on lipid peroxidation by trichloromethyl radicals produced due to metabolism of carbon tetrachloride; acute liver injury that manifests itself in the form of centrilobular necrosis, followed by hepatocyte regeneration[33-35]. Gastric administration can lead to hepatic cirrhosis[36]. |
| 02 | Thioacetamide | Hepatocytes | Acute liver damage by biotransformation into thioacetamide sulfoxide and sulfone causing centrilobular necrosis, followed by hepatocyte regeneration[37,38]. Continued exposure induces hepatic cirrhosis[36]. |
| 03 | Acetaminophen (or paracetamol) | Hepatocytes/Oval cells | Acute liver failure, due to formation of N-acetyl-benzoquinoneimine, resulting in centrilobular apoptosis and necrosis, activation of Kupffer cells, followed by hepatocyte regeneration[39,40]. Exerts its effect only when present in excess and normal detoxifying pathways are saturated[41]. Higher dose leads to oval cell activation in the smallest portal tracts[42]. |
| 04 | D-Galactosamine | Hepatocytes/Oval cells | Acute liver injury by causing intracellular depletion of uridine nucleotides and consequent pan-lobular necrosis, followed by hepatocyte regeneration[43,44]. Increased/repeated dose leads to activation of progenitor or oval cells that proliferate and differentiate into hepatocytes and biliary cells[45,46]. |
| 05 | Ethanol | Hepatocytes/Oval cells | Damage due to hepatocyte steatosis and necrosis, but large variations seen among different studies due to mode and duration of administration[12,47,48]. Causes weakened and delayed liver regeneration after partial hepatectomy[49] |
Besides these, there are different dietary models to induce liver injury and regeneration in mice. 1,4-dihydro-2,4,6-trimethyl-pyridine-3,5-dicarboxylate (DDC)[50] is one such diet, which causes biliary injury and fibrosis[51]. Mice on DDC diet respond poorly to partial hepatectomy[50]. Another commonly used diet is choline-deficient, ethionine-supplemented (CDE) diet, first used in mice by Passman et al.[52], which causes hepatocellular injury with steatosis[53,54]. Though rodents have been the model of choice for liver regeneration studies, the zebrafish has recently been developed as a liver regeneration model that uses surgical partial hepatectomy[55], drug-induced injury[55] and nitroreductase-mediated hepatocyte ablation[56]. The advantages of using zebrafish for liver regeneration studies are low cost, rapid analysis and easy in vivo chemical screening due to the animal’s small size and translucent body, respectively.
Mechanisms of liver regeneration
The restoration of structural and functional hepatic loss due to internal or external injuries can be accomplished by either compensatory hyperplasia of hepatocytes or progenitor/stem cell mediated regeneration[4]. Compensatory hyperplasia takes place in the absence of significant hepatocyte senescence. In such cases, hepatocytes majorly contribute to liver regeneration with very little or no involvement of progenitor/stem cells[16]. On the other hand, if there is a failure of hepatocyte replication or there is severe injury, then hepatic stem/progenitor cells play a major role[15].
Hepatic progenitor cells (HPCs) play a dual role in liver injury, i.e., regeneration and fibrosis[57-59]. HPCs are activated in the periportal area after substantial liver damage. The activated HPCs are then directed towards biliary and hepatic regeneration via up-regulation of the Notch and Wnt-β-catenin pathways, respectively. In addition to regeneration, proliferation of HPCs also leads to fibrosis during liver injury. Different mechanisms are involved in HPC mediated fibrosis and inflammation: Activation of hepatic stellate cells (HSCs) via transforming growth factor (TGF-β) and sonic hedgehog (SHh) signals to produce abnormal extracellular matrix (ECM); recruitment of activated macrophages via chemokine ligand (CCL-2), Tweak, chemokine ligand 5 (CCL5)/RANTES, and intercellular adhesion molecules (ICAMs); stimulation of abnormal angiogenesis by liver endothelial cells[57]. Hence, HPCs participate both in regeneration and fibrogenesis. Also, the presence of progenitor cells and the HPC response have been associated with liver tumor formation[60]. The cellular source for all these functions is provided by the heterogenous cell population in the HPC niche[58,61].
Though various studies have revealed the genes, cytokines, growth factors, and signalling pathways involved in progenitor/oval cell based liver regeneration[62-65], the cellular and molecular mechanisms involved in progenitor/stem cell mediated regeneration are relatively less characterized[4]. Thus, in order to design HPC-based, pro-regenerative and anti-fibrotic therapies, a deeper understanding of the origin of HPCs, their niche components consisting of heterogeneous cell populations, signalling molecules involved in activation, proliferation, migration and differentiation of HPCs, signalling pathways and their relative relevance is required[14,57,58,66-69].
Although the possibility of hepatocyte differentiation from HPCs is not completely excluded, another school of thought also suggests that new hepatocytes and cholangiocytes are derived only from pre-existing hepatocytes and not from HPCs in liver injuries[16,70-72].
Besides these two mechanisms, Nagy et al.[73] (2001) have also shown that if both hepatocyte proliferation and stem cell activation are prevented by dexamethasone or 5-fluorouracil, liver restoration is achieved by hypertrophy/enlargement of periportal hepatocytes.
The whole process of regeneration is completed in three phases, i.e., priming (increased capacity of hepatocytes to replicate), proliferation (attainment of required functional cell mass) and termination (end of proliferation)[5,6]. Each phase is executed by a well-coordinated network of various parenchymal and non-parenchymal cells[74-76], hormones from various glands[7], growth factors, and signalling networks[7,8]. Both intra-hepatic and extra-hepatic cells are involved in liver regeneration, i.e., hepatocytes, sinusoidal endothelial cells (SECs), Kupffer cells, hepatic stellate cells (HSCs), hepatic stem cells, biliary epithelial cells, platelets, eosinophils, platelets, and natural killer T (NKT) cells. Hormones (insulin, glucagon, serotonin, somatostatin, norepinephrine, T3, etc.) from various glands (thyroid, adrenal, pancreas, duodenum), cytokines [tumor necrosis factor (TNF)-α, interleukin [IL]-6, interferon α and γ], transcription factors [NF-κB, STAT3, CCAAT-enhancer binding protein (C/EBP) b, farnesoid X receptor (FXR), cAMP regulatory element-binding protein, activator protein 1] and different growth factors [epidermal growth factor (EGF), transforming growth factor (TGF), hepatocyte growth factor (HGF), vascular endothelial growth factor (VEGF) etc.] also affect the process of regeneration. Optimum liver regeneration results from various signalling mechanisms that are turned on and off at specific times. Some of the prominent pathways involved are: IL-6/Jak/STAT3 pathway in hepatocyte proliferation, PI3-K/PDK1/Akt pathway in hepatocyte growth, HGF/PI3-K/Akt and FXR-p62/SQSTM1 pathways in liver protection etc. An exhaustive list and roles of all these major players in the execution and regulation of liver regeneration has been extensively reviewed elsewhere[7-9,13,77-82]. Besides these cellular and molecular mechanisms, stress, hypoxia, and gut microbiome have also been directly or indirectly linked with liver regeneration, as reviewed by Preziosi et al.[8].
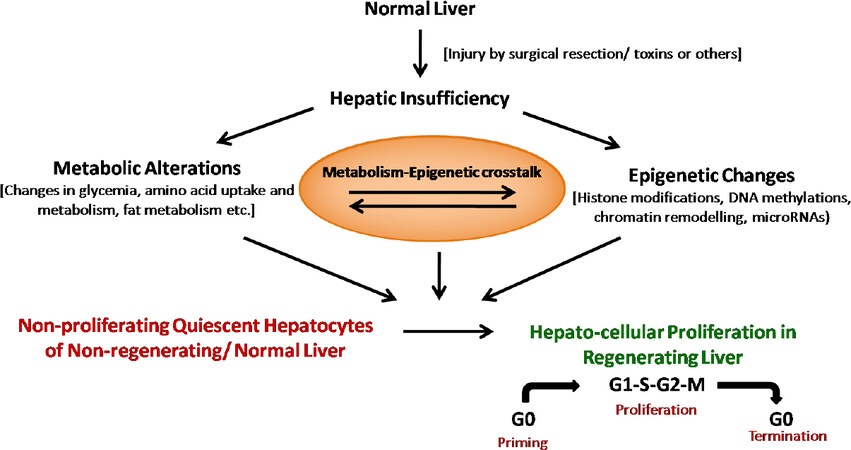
Despite of the tremendous knowledge about these players, we are yet to understand the essence of hepatostat, i.e., the adjustment of liver size to 100% for required homeostasis[83]. Metabolic alterations are one of the early events in hepatic insufficiency, and play an important role in the execution and regulation of liver regeneration[9]. Equally important are the recently reviewed[84-86] epigenetic mechanisms that tightly regulate the expression of genes, specific for each phase of regeneration. Here, we review the potential relationship between metabolism, epigenetics, and liver regeneration.
Metabolic regulation of liver regeneration
Since metabolism is the major function of liver, metabolic alterations are bound to occur in an injured liver. Many studies have shown that both hepatic and systemic metabolism are altered in injury induced liver regeneration. Some of these changes, as reviewed by Huang et al.[9] include: the suppression of liver glycolysis and induction of gluconeogenesis, depletion of glucagon in remnant liver, decline in lean and adipose tissue mass, depletion of systemic fat, steatosis, increase in lipolysis and serum free amino acids, decline in hepatic ATP content and increase in AMP in remnant liver, and β-oxidation of fatty acids as a primary source of ATP production in regenerating liver. These metabolic changes occur before the initiation of surgery or toxin-based hepato-cellular proliferation that is subsequently promoted by cyclin-CDK complexes. The metabolic perturbations get resolved with advancement of regeneration events[87-89].
Various studies have implicated that these changes are not only metabolic perturbations in response to hepatic insufficiency, but physiological determinants of liver regeneration too. For example, the physiological importance of hypoglycaemia is revealed by the fact that glucose supplementation has been shown to impair liver regeneration[87,90]. Similarly, inhibiting liver fat accumulation by using drugs[89,91,92] or knockout mice[88,89] has been shown to suppress liver regeneration. Several other studies have demonstrated that alteration in amino acid uptake and metabolism, which is another metabolic response to hepatic insufficiency, also has physiological relevance in regenerative hepatocellular proliferation[93-97]. Thus, there exists a well-coordinated balance between hepatic insufficiency associated metabolic changes and ensuing regeneration events.
Huang et al.[9] have also suggested various candidate molecular mediators that link metabolism with liver regeneration, though they require definite identification. These molecules probably serve as substrates for energy and synthesis of macromolecules during regeneration. One such class of mediators is xenobiotics, which have been shown to induce hepatocellular hyperplasia and hypertrophy in the absence of liver injury in rodents[98]. Such xenobiotic-induced hepatocellular proliferation is executed by various nuclear receptor transcription factors. Peroxisome proliferator-activated receptor (PPAR)-α is one such nuclear receptor transcription factor, whose expression is required for clofibrate and Wy-14,636 xebobiotic-induced hepatomegaly in rodents[99]. But recently, endogenous lipid metabolites have been reported as ligand activators of PPAR-α[100], which indicates that these naturally occurring ligands might connect the post-hepatectomy lipid accumulation with subsequent hepatocyte proliferation during regeneration. Other such xenobiotic-induced transcription factors, which might link hepatic insufficiency induced altered metabolites with liver regeneration, include constitutive androstane receptor (CAR), famesoid X receptor (FXR) and liver X receptor (LXR)[9].
Metabolism also regulates the signalling molecules and pathways essential for regeneration. For example, epidermal growth factor receptor (EGF-R) ligands regulate hepatocyte proliferation in experimental liver regeneration models[78-80]. EGF-R also plays an important role in lipid and fatty acid metabolism in quiescent and regenerating liver, and also in steatosis in a murine model of non-alcoholic fatty liver diseases (NAFLS)[101-103]. EGF-R expression and activity is inhibited by hyperglycemia in other models[104,105]. Thus, hypoglycaemia induced by partial hepatectomy probably promotes EGF-R signalling and hepatocyte proliferation in liver regeneration. Similar possibility exists for glycogen synthase kinase (GSK)-3 to link hepatic insufficiency induced hypoglycaemia with liver regeneration, as reviewed by Huang et al.[9].
Taken together, these examples suggest that altered metabolism as a result of hepatic insufficiency in an injured liver is physiologically relevant for regulating liver regeneration.
Epigenetic regulation of liver regeneration
Transient, but precisely regulated gene expression is the hallmark of different phases of a liver regeneration event[83]. The transcriptional reprogramming involves early activation of otherwise latent genes of a quiescent liver by transcription factors[106,107], and repression of genes associated with hormone biosynthesis and lipid/ steroid metabolism[108,109]. These transcriptional events are time, cell, and context dependent. Such a tightly controlled gene expression can be accomplished by epigenetic mechanisms, and hence, epigenetic mechanisms must be involved in regulating liver regeneration.
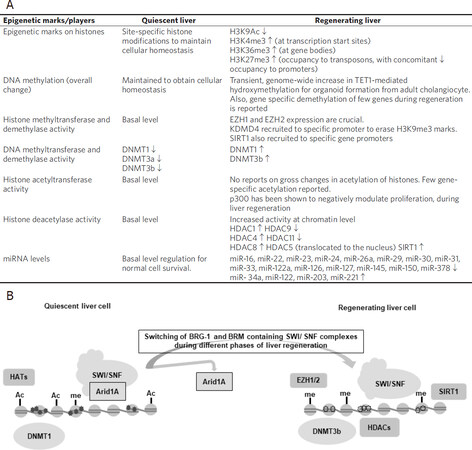
The epigenetic means of transcriptional regulation include DNA methylation, post-translational modifications of histones, microRNA, and chromatin remodelling. So far, only limited reports are available on epigenetic regulation of liver regeneration. Within those, microRNA mediated regulation of liver regeneration has been studied more, which has been reviewed recently[110-112]. Chen et al.[112] have reviewed that miR-16, miR-22, miR-23, miR-24, miR-26a, miR-29, miR-30, miR-31, miR-33, miR-122a, miR-126, miR-127, miR-145 and miR-150, miR-378 are down-regulated; while miR- 34a, miR-122, miR-203, and miR-221 are up-regulated during liver regeneration. miR-21, miR-26b, miR-192 and miR-194 are up-regulated in the first day, and down-regulated in the subsequent two days. miR-21, miR-23b, miR-122, miR-203 and miR-221 are promoters for entry of quiescent hepatocyte into the cell cycle during liver regeneration, whereas miR-26a, miR-33, miR-34a, miR-127, miR-150, and miR-378 are inhibitors for the same.
Among chromatin remodelers, Arid1a, a SWI/SNF (SWItch/Sucrose Non-Fermentable) chromatin remodelling complex component, suppresses hepatocyte proliferation and regeneration[113]. Proliferation of hepatocytes is enhanced in murine liver regeneration models with hepeatocyte-specific deletion of Arid1a. Arid1a limits the access of hepatocyte transcription factors, C/EBPα, hepatocyte nuclear factor α (Hnf4α) and the E2 factor (E2F), to their target genes[113]. Our group has previously reported the switching of Brahma-related gene (BRG-1) and Brahma (BRM) containing SWI/SNF complexes during different phases of thioacetamide induced liver regeneration in mice[114]. This differential expression of SWI/SNF complexes, correlated with histone modification marks, probably regulates the expression of different sets of genes during injury and proliferation phases of liver regeneration.
A cross-talk between DNA methylation and histone modification was recently revealed by Wang et al.[115] in liver regeneration. A dynamic expression of UHRF1, an epigenetic regulator for DNA methylation, was observed during liver regeneration. Also, they found an early and sustained activation of pro-regenerative genes and thus enhanced liver regeneration in partially hepatectomized UHRF1 deleted livers. In these organisms, H3K27me3, a marker for transcriptional repression, was redistributed from promoters to transposones, thus allowing the expression of liver regeneration specific genes.
Bromodomains of bromodomain and extraterminal (BET) proteins, which regulate transcription by binding to acetylated lysine residues on histone tails, also regulate hepatocellular proliferation in hepatocyte-driven liver regeneration[116]. Russell et al.[116] have shown that the drug JQ1, a specific inhibitor of Brd4 (one of BET proteins), impairs hepatocyte proliferation in partial hepatectomy and acetaminophen-induced liver regeneration with significant reduction in E2f2 genes and cyclin-D1.
A couple of studies have shown that p300 histone acetyltransferase (HAT) along with C/EBP transcription factor also regulates hepatocyte proliferation[117-119]. Hepatocellular proliferation gets suppressed when a broad inhibitor of histone deacetyalses (HDACs), suberoylanilide hydroxamic acid (SAHA), is administered in rodents subjected to partial hepatectomy[120]. An absence of enhancer of zeste homologs (EZH) 1 and 2, the methyltransferases, causes loss of regeneration capacity in CCl4 and partial hepatectomy induced liver regeneration. Partly, this is attributed to a decrease in H3K27me3 levels on the promoters of genes that encode G1/S phase cell cycle inhibitors like CDKN2A and CDKN2B[121].
Recently, the highly conserved Hippo pathway and its downstream effectors, the transcriptional co-activators, yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ or WWTR1), have been shown to mediate liver regeneration[122-125] by activating hepatocyte proliferation and trans-differentiation into stem cell like progenitor cells[126]. Interestingly, Hippo signalling is, in turn, regulated by lysine demethylase 3A (KDM3A), the enzyme responsible for removing H3K9me2 and recruiting p300[127]. Also, the EZH2- H3K27me3-DNMT1 complex regulates the wwc1 gene, a key upstream factor of the Hippo pathway[128]. Thus, Hippo signalling, which plays an important role in regeneration, is also regulated by epigenetic mechanisms. Aloia et al.[129] have also shown that epigenetic remodelling through Ten-eleven translocation (TET1)-mediated hydroxymethylation, licences adult cholangiocytes to initiate organoids and activate the liver regeneration through transcriptional regulation of stem-cell genes and regenerative pathways, including YAP-Hippo signalling.
This information, summarized in Figure 1, highlights the regulatory role that epigenetic mechanisms exert during liver regeneration. However, phase and cell-specific studies will help unravel the regulatory role of epigenetic mechanisms in liver regeneration via orchestration of transcriptional reprogramming.
Figure 1. Summary of epigenetic modifications associated with liver regeneration[84,86,112,126-129]. A: Table representing the comparison of a quiescent and regenerating liver. “↑” represents an increase in expression and “↓” represents a decrease in expression; B: A schematic representation of the changes associated with the transition of a quiescent liver to a regeneration phase. During regeneration, a reorganization of histone modifications (primarily, acetylation, and methylation) and DNA methylation occurs. However, a detailed epigenetic code for liver regeneration is yet to be decoded
Metabolites as epigenetic modifiers: an interplay
Hepatic insufficiency due to liver injury causes metabolic perturbations, which are physiologically relevant for liver regeneration. The regeneration event is executed by tightly regulated and phase-specific gene expression that is under epigenetic control. Hence, we hypothesize the existence of a metabolic-epigenetic-liver regeneration link.
Epigenetic mechanisms control transcription with the help of various enzymes like DNA methyltransferases (DNMT), Histone acetyltransferases (HAT), Histone deacetylases (HDAC), Histone methyltransferases (HMT), etc. The activity of these enzymes in turn depends on the availability of their substrates or cofactors, which are nothing but cellular intermediary metabolites. For example, acetyl-CoA, a metabolite, is required for histone acetylation by HAT enzymes, NAD+ is required by sirtuins (class III HDACs) for histone deacetylation, S-adenosylmethionine (SAM) is required for histone/DNA methyltransferases for methylation, FAD+ is needed for demethylases, and so on[130]. Thus, a coupling exists between the metabolic state of a cell and the chromatin-dependent gene regulation[131-134].
Here, we cite certain examples which support the idea of existence of a metabolic-epigenetic-liver regeneration link.
Metabolic alteration regulates DNA methylation during liver regeneration
Mato and Lu (2007)[135] have shown that the synthesis of S-adenosyl-methionine (SAMe) by methionine-adenosyl transferase 1A (MAT1A) from methionine remains tightly regulated during liver regeneration, and gets disrupted in liver diseases including cancer. An altered expression of MAT1A disrupts methionine metabolism, which has been shown to inhibit liver regeneration in mice[93]. Since SAMe is required for methyltransferases’ activity, DNA methylation might be altered in such cases. DNA methylation, in turn, regulates liver regeneration, similar to what has been demonstrated in another study, wherein, azacytidine, the methyltransferase inhibitor, suppresses liver regeneration[136].
Another example linking metabolism and DNA methylation during liver regeneration comes from α-ketoglutarate (α-KG). α-KG is an amino group acceptor in an alanine aminotransferase catalyzed reaction for the production of a gluconeogenic precursor, pyruvate, from alanine. The metabolic alteration in levels of α-KG is likely to occur along with glycolytic and gluconeogenic changes in a regenerating liver. α-KG is also the co-factor for demethylation reactions[137]. Also, the TET-1 catalytic activity required for cholangiocyte organoid initiation and maintenance depends on α-KG[129,138]. Hence, metabolic changes in α-KG might alter DNA methylation and ensuing gene expression patterns in a regenerating liver[9].
Metabolic alteration regulates histone acetylation during liver regeneration
Parallel to hypoglycaemia, zinc-dependent HDAC activity increases and global liver histone acetylation decreases in liver regeneration. SAHA, the HDAC inhibitor, is reported to suppress liver regeneration[139]. Thus, glycemia seems to affect liver regeneration by regulating protein acetylation level. Also, the sub-cellular localization of HDAC5, a class-IIa zinc-dependent HDAC, is also regulated by glycemia. In partial hepatectomy, the glycemic alteration brings about this localization in the nucleus[140].
Shimazu et al.[141] have reported that β-hydroxybutyrate, a ketone body, is a specific and endogenous inhibitor of class-I HDACs, supporting the idea of coupling between epigenetic transcriptional regulation and metabolic status of a cell. There is a possibility that other metabolites generated during hepatic insufficiency in liver regeneration might also activate/inhibit the epigenetic players. A possible link between glucose homeostasis and fat metabolism with the SIRT1, a class-III HDAC, has been reported during liver regeneration[142].
These examples indicate that a metabolic-epigenetic-liver regeneration link exists that helps in the well-regulated execution of molecular events during liver regeneration as depicted below in Figure 2.
Figure 2. Hepatic insufficiency caused by surgical or toxin-induced injury induces metabolic alterations and epigenetic changes in the liver. Besides the isolated metabolic and epigenetic regulation of hepatocyte proliferation and regeneration, a well-controlled interplay between metabolism and epigenetics controls liver regeneration events
Summary and conclusions
Hepatic insufficiency caused by liver injury generates altered metabolites. These metabolites are physiologically relevant for hepatocellular proliferation and regeneration. These metabolites also supply the substrates/cofactors for various enzymes involved in epigenetic-mediated transcriptional regulation. Though an increasing number of isolated reports are available for metabolic and epigenetic regulation of liver regeneration, studies focusing on the inter-dependence between metabolism and epigenetics during liver regeneration need to be investigated. Investigation of the metabolic-epigenetic interplay and its molecular characterization during liver regeneration can provide better insights in identifying potential targets for clinical treatments in liver related diseases.
Declarations
Authors’ contributionsWrote the major part of the manuscript: Verma S
Helped in writing the manuscript and made the epigenetic model: Purohit JS
Helped in material collection and reference arrangement: Arora A, Sinha S
Envisaged the concept, designed an outlines, and corrected the manuscript: Chaturvedi MM
Availability of data and materialsNot applicable.
Financial support and sponsorshipSV acknowledges the support received from DDU College, University of Delhi; AA acknowledges Maitreyi College (University of Delhi); JSP and MMC acknowledge the Departments of Science and Technology, India and University of Delhi for their financial support in pursuing the research work presented in this manuscript.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
1. Sepanlou SG, Safiri S, Bisignano C, et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2020;5:245-66.
2. Global burden of liver disease: a true burden on health sciences and economies!! World Gastroenterology Organisation. Available from: https://www.worldgastroenterology.org/publications/e-wgn/e-wgn-expert-point-of-view-articles-collection/global-burden-of-liver-disease-a-true-burden-on-health-sciences-and-economies.[Last accessed on 2 Oct 2020].
3. Ueno S, Sakoda M, Kubo F, et al; Kagoshima Liver Cancer Study Group. Surgical resection versus radiofrequency ablation for small hepatocellular carcinomas within the Milan criteria. J Hepatobiliary Pancreat Surg 2009;16:359-66.
4. Kholodenko IV, Yarygin KN. Cellular mechanisms of liver regeneration and cell-based therapies of liver diseases. Biomed Res Int 2017;2017:8910821.
6. Court FG, Wemyss-Holden SA, Dennison AR, Maddern GJ. The mystery of liver regeneration. Br J Surg 2002;89:1089-95.
7. Ozaki M. Cellular and molecular mechanisms of liver regeneration: proliferation, growth, death and protection of hepatocytes. Semin Cell Dev Biol 2020;100:62-73.
8. Preziosi ME, Monga SP. Update on the Mechanisms of Liver Regeneration. Semin Liver Dis 2017;37:141-51.
9. Huang J, Rudnick DA. Elucidating the metabolic regulation of liver regeneration. Am J Pathol 2014;184:309-21.
10. Hu H, Gehart H, Artegiani B, et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 2018;175:1591-1606.e19.
11. Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299-312.
13. Forbes SJ, Newsome PN. Liver regeneration - mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473-85.
14. Santoni-Rugiu E, Jelnes P, Thorgeirsson SS, Bisgaard HC. Progenitor cells in liver regeneration: molecular responses controlling their activation and expansion. APMIS 2005;113:876-902.
15. Lu WY, Bird TG, Boulter L, et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 2015;17:971-83.
16. Yanger K, Knigin D, Zong Y, et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014;15:340-9.
17. Experimental pathology of liver: restoration of liver in white rat following partial surgical removal. ScienceOpen. Available from: https://www.scienceopen.com/document?vid=57858414-5eff-4c8d-a028-ccb3fc1c44a6. [Last accessed on 30 Sep 2020].
18. Sakamoto T, Ezure T, Lunz J, et al. Concanavalin A simultaneously primes liver hematopoietic and epithelial progenitor cells for parallel expansion during liver regeneration after partial hepatectomy in mice. Hepatology 2000;32:256-67.
19. Ku Y, Tominaga M, Sugimoto T, et al. Preoperative hepatic venous embolization for partial hepatectomy combined with segmental resection of major hepatic vein. Br J Surg 2002;89:63-9.
20. Kahn D, Hickman R, Terblanche J. A porcine model for the study of liver regeneration. J Invest Surg 1988;1:139-42.
21. Kahn D, Hickman R, Terblanche J, von Sommoggy S. Partial hepatectomy and liver regeneration in pigs—The response to different resection sizes. J Surg Res 1988;45:176-80.
22. Gaglio PJ, Liu H, Dash S, et al. Liver regeneration investigated in a non-human primate model (Macaca mulatta). J Hepatol 2002;37:625-32.
23. Rozga J, Jeppsson B, Bengmark S. Portal branch ligation in the rat. Reevaluation of a model. Am J Pathol 1986;125:300-308.
24. Iwakiri Y, Cadelina G, Sessa WC, Groszmann RJ. Mice with targeted deletion of eNOS develop hyperdynamic circulation associated with portal hypertension. Am J Physiol Gastrointest Liver Physiol 2002;283:G1074-81.
25. Ueno S, Aikou T, Tanabe G, et al. Exogenous hepatocyte growth factor markedly stimulates liver regeneration following portal branch ligation in dogs. Cancer Chemother Pharmacol 1996;38:233-7.
26. Krupski G, Bröring DC, Wittkugel O, et al. Portale Kollateralenbildung nach Pfortaderligatur zur Leberregenerations-Induktion im Schweine-Modell [Formation of portal venous collaterals after ligation of the portal vein for induction of liver regeneration]. Rofo 2002;174:1281-4.
27. Nishida M, Yano K, Murakami T, Suzuki T. Introduction of monoclonal antibodies to bromodeoxyuridine to monitor hepatic regeneration. Gastroenterology 1991;100:1135-7.
28. The effect of age on regeneration of rat liver. Cancer Research. Available at: https://cancerres.aacrjournals.org/content/10/5/324. [Accessed September 30, 2020].
29. Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 2008;3:1167-70.
30. Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 2003;302:255-9.
31. Diehl AM. Nutrition, hormones, metabolism, and liver regeneration. Semin Liver Dis 1991;11:315-20.
32. Yan M, Huo Y, Yin S, Hu H. Mechanisms of acetaminophen-induced liver injury and its implications for therapeutic interventions. Redox Biol 2018;17:274-83.
33. Das P, Chopra P, Nayak N. Hepatocellular tolerance to carbon tetrachloride induced injury in the rat: A study of its nature and possible mode of evolution. Exp Mol Pathol 1974;21:218-36.
34. Wong FW, Chan WY, Lee SS. Resistance to carbon tetrachloride-induced hepatotoxicity in mice which lack CYP2E1 expression. Toxicol Appl Pharmacol 1998;153:109-18.
35. Weber LW, Boll M, Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit Rev Toxicol 2003;33:105-36.
36. Dashti H, Jeppsson B, Hägerstrand I, et al. Thioacetamide- and carbon tetrachloride-induced liver cirrhosis. Eur Surg Res 1989;21:83-91.
37. Chieli E, Malvaldi G. Role of the microsomal fad-containing monooxygenase in the liver toxicity of thioacetamide S-oxide. Toxicology 1984;31:41-52.
38. Mangipudy RS, Chanda S, Mehendale HM. Hepatocellular regeneration: key to thioacetamide autoprotection. Pharmacol Toxicol 1995;77:182-8.
40. Zieve L, Anderson WR, Dozeman R, Draves K, Lyftogt C. Acetaminophen liver injury: sequential changes in two biochemical indices of regeneration and their relationship to histologic alterations. J Lab Clin Med 1985;105:619-624.
41. Tuñón MJ, Alvarez M, Culebras JM, González-Gallego J. An overview of animal models for investigating the pathogenesis and therapeutic strategies in acute hepatic failure. World J Gastroenterol 2009;15:3086-98.
42. Kofman AV, Morgan G, Kirschenbaum A, et al. Dose- and time-dependent oval cell reaction in acetaminophen-induced murine liver injury. Hepatology 2005;41:1252-61.
43. El-Mofty SK, Scrutton MC, Serroni A, Nicolini C, Farber JL. Early, reversible plasma membrane injury in galactosamine-induced liver cell death. Am J Pathol 1975;79:579-96.
44. Macdonald JR, Thayer KJ, White C. Inhibition of galactosamine cytotoxicity in an in vivo/in vitro hepatocellular toxicity model. Toxicol Appl Pharmacol 1987;89:269-77.
45. Dabeva MD, Shafritz DA. Activation, proliferation, and differentiation of progenitor cells into hepatocytes in the D-galactosamine model of liver regeneration. Am J Pathol 1993;143:1606-20.
46. Lemire JM, et al. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. Am J Pathol 1991;139:535-52.
47. Frank WO, et al. Effect of acute ethanol administration upon hepatic regeneration. J Lab Clin Med 1979;93:402-13.
48. Morales-González JA, Gutiérrez-Salinas J, Yáñez L, Villagómez-Rico C, Badillo-Romero J, Hernández-Muñoz R. Morphological and biochemical effects of a low ethanol dose on rat liver regeneration: role of route and timing of administration. Dig Dis Sci 1999;44:1963-74.
49. Akerman PA, Cote PM, Yang SQ, et al. Long-term ethanol consumption alters the hepatic response to the regenerative effects of tumor necrosis factor-alpha. Hepatology 1993;17:1066-73.
50. Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest 1999;79:103-9.
51. Williams MJ, Clouston AD, Forbes SJ. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology 2014;146:349-56.
52. Passman AM, Strauss RP, McSpadden SB, et al. A modified choline-deficient, ethionine-supplemented diet reduces morbidity and retains a liver progenitor cell response in mice. Dis Model Mech 2015;8:1635-41.
53. Boulter L, Govaere O, Bird TG, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 2012;18:572-9.
54. Tsuchiya A, Lu WY, Weinhold B, et al. Polysialic acid/neural cell adhesion molecule modulates the formation of ductular reactions in liver injury. Hepatology 2014;60:1727-40.
55. Goessling W. Liver Regeneration in Zebrafish. Liver Regeneration. Elsevier; 2015. pp. 41-7.
56. Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc 2008;3:948-54.
57. Kaur S, Siddiqui H, Bhat MH. Hepatic progenitor cells in action: liver regeneration or fibrosis? Am J Pathol 2015;185:2342-50.
58. Lukacs-Kornek V, Lammert F. The progenitor cell dilemma: cellular and functional heterogeneity in assistance or escalation of liver injury. J Hepatol 2017;66:619-30.
59. Greenbaum LE, Wells RG. The role of stem cells in liver repair and fibrosis. Int J Biochem Cell Biol 2011;43:222-9.
60. Libbrecht L. Hepatic progenitor cells in human liver tumor development. World J Gastroenterol 2006;12:6261-5.
62. Batusic DS, Cimica V, Chen Y, et al. Identification of genes specific to “oval cells” in the rat 2-acetylaminofluorene/partial hepatectomy model. Histochem Cell Biol 2005;124:245-60.
63. Cimica V, Batusic D, Chen Y, Hollemann T, Pieler T, Ramadori G. Transcriptome analysis of rat liver regeneration in a model of oval hepatic stem cells. Genomics 2005;86:352-64.
64. Sánchez A, Factor VM, Schroeder IS, Nagy P, Thorgeirsson SS. Activation of NF-kappaB and STAT3 in rat oval cells during 2-acetylaminofluorene/partial hepatectomy-induced liver regeneration. Hepatology 2004;39:376-85.
65. Lowes KN, Croager EJ, Olynyk JK, Abraham LJ, Yeoh GC. Oval cell-mediated liver regeneration: Role of cytokines and growth factors. J Gastroenterol Hepatol 2003;18:4-12.
66. Chen J, Chen L, Zern MA, et al. The diversity and plasticity of adult hepatic progenitor cells and their niche. Liver Int 2017;37:1260-71.
67. Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 2014;14:561-74.
68. Van Haele M, Roskams T. Hepatic Progenitor Cells: An Update. Gastroenterol Clin North Am 2017;46:409-20.
69. Vestentoft PS. Development and molecular composition of the hepatic progenitor cell niche. Dan Med J 2013;60:B4640.
70. Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep 2014;8:933-9.
71. Tarlow BD, Finegold MJ, Grompe M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology 2014;60:278-89.
72. Sekiya S, Suzuki A. Hepatocytes, rather than cholangiocytes, can be the major source of primitive ductules in the chronically injured mouse liver. Am J Pathol 2014;184:1468-78.
73. Nagy P, Teramoto T, Factor VM, et al. Reconstitution of liver mass via cellular hypertrophy in the rat. Hepatology 2001;33:339-45.
74. Fujiyoshi M, Ozaki M. Molecular mechanisms of liver regeneration and protection for treatment of liver dysfunction and diseases. J Hepatobiliary Pancreat Sci 2011;18:13-22.
76. Pahlavan PS, Feldmann RE Jr, Zavos C, Kountouras J. Prometheus’ challenge: molecular, cellular and systemic aspects of liver regeneration. J Surg Res 2006;134:238-51.
79. Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol 2010;176:2-13.
81. Batusic DS, von Bargen A, Blaschke S, Dudas J, Ramadori G. Different physiology of interferon-α/-γ in models of liver regeneration in the rat. Histochem Cell Biol 2011;136:131-44.
82. Hoffmann K, Nagel AJ, Tanabe K, et al. Markers of liver regeneration-the role of growth factors and cytokines: a systematic review. BMC Surg 2020;20:31.
83. Michalopoulos GK. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology 2017;65:1384-92.
84. Arechederra M, Berasain C, Avila MA, Fernández-Barrena MG. Chromatin dynamics during liver regeneration. Semin Cell Dev Biol 2020;97:38-46.
85. Moran-Salvador E, Mann J. Epigenetics and Liver Fibrosis. Cell Mol Gastroenterol Hepatol 2017;4:125-34.
86. Monga SP, Sadler KC. An epigenetic perspective on liver regeneration. Epigenomics 2020;12:381-4.
87. Weymann A, Hartman E, Gazit V, et al. p21 is required for dextrose-mediated inhibition of mouse liver regeneration. Hepatology 2009;50:207-15.
88. Gazit V, Weymann A, Hartman E, et al. Liver regeneration is impaired in lipodystrophic fatty liver dystrophy mice. Hepatology 2010;52:2109-17.
89. Shteyer E, Liao Y, Muglia LJ, Hruz PW, Rudnick DA. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology 2004;40:1322-32.
90. Holeček M. Nutritional modulation of liver regeneration by carbohydrates, lipids, and amino acids: a review. Nutrition 1999;15:784-8.
91. Srinivasan SR, Chow CK, Glauert HP. Effect of the peroxisome proliferator ciprofibrate on hepatic DNA synthesis and hepatic composition following partial hepatectomy in rats. Toxicology 1990;62:321-32.
92. Walldorf J, Hillebrand C, Aurich H, et al. Propranolol impairs liver regeneration after partial hepatectomy in C57Bl/6-mice by transient attenuation of hepatic lipid accumulation and increased apoptosis. Scand J Gastroenterol 2010;45:468-76.
93. Chen L, Zeng Y, Yang H, et al. Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J 2004;18:914-6.
94. Brebnor LD, Balinsky JB. Changes in activities of urea cycle enzymes in early stages of liver regeneration after partial hepatectomy in rats. Life Sci 1983;32:1391-400.
95. Gebhardt R. Altered acinar distribution of glutamine synthetase and different growth response of cultured enzyme-positive and -negative hepatocytes after partial hepatectomy. Cancer Res 1990;50:4407-10.
96. Freeman TL, Ngo HQ, Mailliard ME. Inhibition of system A amino acid transport and hepatocyte proliferation following partial hepatectomy in the rat. Hepatology 1999;30:437-44.
97. Holecek M. Three targets of branched-chain amino acid supplementation in the treatment of liver disease. Nutrition 2010;26:482-90.
98. Hall AP, Elcombe CR, Foster JR, et al. Liver hypertrophy: a review of adaptive (adverse and non-adverse) changes--conclusions from the 3rd International ESTP Expert Workshop. Toxicol Pathol 2012;40:971-94.
99. Lee SS, Pineau T, Drago J, et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol 1995;15:3012-22.
100. Chakravarthy MV, Lodhi IJ, Yin L, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell 2009;138:476-88.
101. Bhushan B, Michalopoulos GK. Role of epidermal growth factor receptor in liver injury and lipid metabolism: Emerging new roles for an old receptor. Chem Biol Interact 2020;324:109090.
102. López-Luque J, Caballero-Díaz D, Martinez-Palacián A, et al. Dissecting the role of epidermal growth factor receptor catalytic activity during liver regeneration and hepatocarcinogenesis. Hepatology 2016;63:604-19.
103. Bhushan B, Banerjee S, Paranjpe S, et al. Pharmacologic inhibition of epidermal growth factor receptor suppresses nonalcoholic fatty liver disease in a murine fast-food diet model. Hepatology 2019;70:1546-63.
104. Obata T, Maegawa H, Kashiwagi A, Pillay TS, Kikkawa R. High glucose-induced abnormal epidermal growth factor signaling. J Biochem 1998;123:813-20.
105. Blackshear PJ, Stumpo DJ, Kennington EA, et al. Decreased levels of hepatic epidermal growth factor receptors in obese hyperglycemic rodents. J Biol Chem 1987;262:12356-64.
106. Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci U S A 2002;99:11181-6.
107. Kelley-Loughnane N, Sabla GE, Ley-Ebert C, Aronow BJ, Bezerra JA. Independent and overlapping transcriptional activation during liver development and regeneration in mice. Hepatology 2002;35:525-34.
108. White P, Brestelli JE, Kaestner KH, Greenbaum LE. Identification of transcriptional networks during liver regeneration. J Biol Chem 2005;280:3715-22.
109. Li J, Campbell JS, Mitchell C, et al. Relationships between deficits in tissue mass and transcriptional programs after partial hepatectomy in mice. Am J Pathol 2009;175:947-57.
110. Yi P, Zhang M, Xu M. Role of microRNA in liver regeneration. Hepatobiliary Pancreat Dis Int 2016;15:141-6.
111. Lauschke VM, Mkrtchian S, Ingelman-Sundberg M. The role of microRNAs in liver injury at the crossroad between hepatic cell death and regeneration. Biochem Biophys Res Commun 2017;482:399-407.
112. Chen X, Zhao Y, Wang F, Bei Y, Xiao J, Yang C. MicroRNAs in Liver Regeneration. Cell Physiol Biochem 2015;37:615-28.
113. Sun X, Chuang JC, Kanchwala M, et al. Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell 2016;18:456-66.
114. Sinha S, Verma S, Chaturvedi MM. Differential expression of SWI/SNF chromatin remodeler subunits Brahma and Brahma-Related gene during drug-induced liver injury and regeneration in mouse model. DNA Cell Biol 2016;35:373-84.
115. Wang S, Zhang C, Hasson D, et al. Epigenetic compensation promotes liver regeneration. Dev Cell 2019;50:43-56.e6.
116. Russell JO, Ko S, Saggi HS, et al. Bromodomain and extraterminal (BET) proteins regulate hepatocyte proliferation in hepatocyte-driven liver regeneration. Am J Pathol 2018;188:1389-405.
117. Bricambert J, Miranda J, Benhamed F, Girard J, Postic C, Dentin R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest 2010;120:4316-31.
118. Fang S, Tsang S, Jones R, et al. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem 2008;283:35086-95.
119. Breaux M, Lewis K, Valanejad L, et al. p300 regulates liver functions by controlling p53 and C/EBP family proteins through multiple signaling pathways. Mol Cell Biol 2015;35:3005-16.
120. Huang J, Schriefer AE, Yang W, Cliften PF, Rudnick DA. Identification of an epigenetic signature of early mouse liver regeneration that is disrupted by Zn-HDAC inhibition. Epigenetics 2014;9:1521-31.
121. Bae WK, Kang K, Yu JH, et al. The methyltransferases enhancer of zeste homolog (EZH) 1 and EZH2 control hepatocyte homeostasis and regeneration. FASEB J 2015;29:1653-62.
122. Loforese G, Malinka T, Keogh A, et al. Impaired liver regeneration in aged mice can be rescued by silencing Hippo core kinases MST1 and MST2. EMBO Mol Med 2017;9:46-60.
123. Lu L, Finegold MJ, Johnson RL. Hippo pathway coactivators Yap and Taz are required to coordinate mammalian liver regeneration. Exp Mol Med 2018;50:e423.
124. Grijalva JL, Huizenga M, Mueller K, et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am J Physiol Gastrointest Liver Physiol 2014;307:G196-204.
125. Bai H, Zhang N, Xu Y, et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology 2012;56:1097-107.
126. Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 2019;20:211-26.
127. Wang HY, Long QY, Tang SB, et al. Histone demethylase KDM3A is required for enhancer activation of hippo target genes in colorectal cancer. Nucleic Acids Res 2019;47:2349-64.
128. Liu X, Li C, Zhang R, et al. The EZH2- H3K27me3-DNMT1 complex orchestrates epigenetic silencing of the wwc1 gene, a Hippo/YAP pathway upstream effector, in breast cancer epithelial cells. Cell Signal 2018;51:243-56.
129. Aloia L, McKie MA, Vernaz G, et al. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat Cell Biol 2019;21:1321-33.
130. Etchegaray JP, Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell 2016;62:695-711.
132. Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature 2013;502:489-98.
133. Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol 2015;10:95-108.
134. Tzika E, Dreker T, Imhof A. Epigenetics and metabolism in health and disease. Front Genet 2018;9:361.
135. Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology 2007;45:1306-12.
136. Cihák A, Seifertová M, Veselý J, Sorm F. Metabolic alterations of liver regeneration. 8. Enhanced synthesis of DNA in the liver of 5-azacytidine-treated rats subjected to partial hepatectomy. Int J Cancer 1972;10:20-7.
138. Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011;333:1300-3.
139. Huang J, Barr E, Rudnick DA. Characterization of the regulation and function of zinc-dependent histone deacetylases during rodent liver regeneration. Hepatology 2013;57:1742-51.
140. Mihaylova MM, Vasquez DS, Ravnskjaer K, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 2011;145:607-21.
141. Shimazu T, Hirschey MD, Newman J, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013;339:211-4.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Verma S, Purohit JS, Arora A, Sinha S, Chaturvedi MM. Liver regeneration: metabolic and epigenetic regulation. Hepatoma Res 2021;7:16. http://dx.doi.org/10.20517/2394-5079.2020.122
AMA Style
Verma S, Purohit JS, Arora A, Sinha S, Chaturvedi MM. Liver regeneration: metabolic and epigenetic regulation. Hepatoma Research. 2021; 7: 16. http://dx.doi.org/10.20517/2394-5079.2020.122
Chicago/Turabian Style
Verma, Sudhir, Jogeswar S. Purohit, Anshu Arora, Sonal Sinha, Madan Mohan Chaturvedi. 2021. "Liver regeneration: metabolic and epigenetic regulation" Hepatoma Research. 7: 16. http://dx.doi.org/10.20517/2394-5079.2020.122
ACS Style
Verma, S.; Purohit JS.; Arora A.; Sinha S.; Chaturvedi MM. Liver regeneration: metabolic and epigenetic regulation. Hepatoma. Res. 2021, 7, 16. http://dx.doi.org/10.20517/2394-5079.2020.122
About This Article
Copyright
Data & Comments
Data

 Cite This Article 28 clicks
Cite This Article 28 clicks










Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.