Expression quantitative trait loci for PVT1 contributes to the prognosis of hepatocellular carcinoma
Abstract
Aim: Plasmacytoma variant translocation 1 (PVT1), a long intergenic non-coding RNA, was overexpressed in liver cancer. A single nucleotide polymorphism (SNP) rs4733586 was identified as an expression quantitative trait loci (eQTL) for PVT1 using bioinformatics analysis. This study was to assess the association of PVT1 eQTL with hepatocellular carcinoma (HCC) prognosis.
Methods: A case-only study was performed to assess the association between SNP and HCC overall survival in 331 HCC patients with hepatitis B virus. Cox proportional hazard regression models were conducted for survival analysis with adjustment for age, gender, smoking status, drinking status, Barcelona-Clinic Liver Cancer stages, and chemotherapy or transcatheter hepatic arterial chemoembolization (TACE) status.
Results: The variant genotype C allele of rs4733586 was significantly associated with a higher death risk compared with T allele (adjusted hazard ratio = 1.26, 95% confidence intervals = 1.05-1.51, P = 0.012 in the additive model). By stepwise Cox proportional hazard analysis, four variables (age, drinking status, chemotherapy or TACE status, PVT1 eQTL) were remained in the final regression model. In the stratified analysis, no heterogeneity was observed among different subgroups.
Conclusion: These findings suggest that eQTL SNP for PVT1 may be susceptibility marker for the HCC overall survival.
Keywords
Introduction
Liver cancer is the second most common cause of cancer death in the world, of which hepatocellular carcinoma (HCC) is the predominant form[1]. Ranked as the sixth most common form of cancer, HCC is also the third leading cause of cancer death[2]. In previous study, 3-year survival rate among patients at intermediate stages was 50%, whereas among those at advanced stage was just 8%[3]. Although several therapies including radiofrequency ablation, liver transplantation, tumor resection and some others are the potentially effective treatments for HCC, HCC still has a poor 5-year survival rate of about 7%[4,5]. Due to different factors of disease and the poor survival outcomes of HCC patients, it is crucial to identify beneficial molecular biomarker to guide individualized treatment and to improve the prognosis of cancer patients.
Non-coding RNAs (ncRNAs) are emerging as novel regulatory factor in the cancer paradigm[6]. Long noncoding RNAs (lncRNAs), longer than 200 nucleotides in length, are evolutionarily conserved non-protein coding RNAs[7]. LncRNAs have been reported to play an important role in various biological processes related to cancer progressions, such as proliferation, apoptosis and invasion. Plasmacytoma variant translocation 1 (PVT1), a long intergenic non-coding RNA, is located in the chr8q24.21 region[7]. Chromosome 8q24 contains a locus conferring an increased risk for multiple cancers[8]. Recently, several studies have found that PVT1 was functioned as an oncogene and was overexpressed in human tumors including cervical cancer, serous melanoma and prostate cancer[9]. In addition, it was also reported that PVT1 overexpression was associated with clinicopathological features and reduced patients’ survival times[9]. However, the potential function of PVT1 expression quantitative trait loci (eQTL) in the prognosis of HCC has been rarely discussed.
In this study, we identified one single nucleotide polymorphism (SNP) (rs4733586) that may be the eQTL for PVT1 (http://www.regulomedb.org) by using the bioinformatics analysis. Therefore, we thought that the SNP rs4733586 may be likely to regulate the expression of PVT1. Here, we assumed that PVT1 eQTL may contribute to the development and progression of HCC. To verify our hypothesis, we examined the effect of the PVT1 eQTL (rs4733586) on the HCC prognosis of 331 patients from Han population.
Methods
Study subjects
This study was authorized by the local institutional review board at Nanjing Medical University. After approval by the ethics committees, all the participants were given written informed consent, and the registration of the participants was described before[10,11]. In brief, all the patients were consecutively recruited from Nantong Tumor Hospital and the First Affiliated Hospital of Nanjing Medical University, Jiangsu, China[12], and were face-to-face interviewed to collect the demographic information including age, gender, smoking and drinking status. We recruited patients with HCC with hepatitis B virus (HBV) and excluded those with hepatitis C virus (HCV). All the subjects were diagnosed as HCC by histopathological examination. To construct a relatively homogeneous population, our study was limited to HCC patients who have not undergone surgery in intermediate stage (B) or advanced stage (C) according to the Barcelona Clinic Liver Cancer (BCLC) staging system[13]. Eventually, 331 of 414 intermediate or advanced HCC patients completed the follow-ups with the response rate of 80.0% and were performed the survival analysis. We followed up the study subjects every 3 months from the time of recruitment until the death or the last time of follow-up (January 2013).
Serological testing
As described in previous study[11], HBsAg, anti-HBs, anti-HBc and anti-HCV were detected from every patient’s collected serum by following the step of the enzyme-linked immunosorbent assay (Kehua Bio-engineering Co., Ltd., Shanghai, China).
SNP selection and genotyping
We found one common eQTL SNP (rs4733586) in the intron region of lncRNA PVT1 based on the criteria of minor allele frequency (MAF) > 0.05 in Han Chinese from Regulome database. The genomic DNA was extracted from the leukocyte pellet by a series of treatments using conventional methods[14]. Then, we use the Sequenom Mass ARRAY iPLEX platform (Sequenom Inc) to genotype the SNP rs4733586. The information of primers was shown in Table 1. To reduce the false positive rates and error rates, three blank (water) controls were detected in each 384-well plate during samples testing every time. To controlling the quality and yield a 100% concordance rate, more than 10% samples were randomly selected to repeat.
Information of primers for Sequenom MassARRAY iPLEX
| SNP | Primer | Sequence (5’-3’) |
|---|---|---|
| rs4733586 | 2nd-PCR Primer | ACGTTGGATGCAGATTGGAGAGTAGTGGCT |
| 1st-PCR Primer | ACGTTGGATGACATCCGCCCTGGGTGATTC | |
| Extend Primer | GTAGTGGCTCATCACA |
Statistical analysis
We calculated the median survival time (MST), and if the MST could not be calculated, then we use the mean survival time instead. Univariate and multivariable Cox proportional hazard regression analysis was performed to estimate the crude or adjusted hazard ratio (HR) and their 95% confidence intervals (CI), with adjustment of age, gender, smoking status, drinking status, BCLC stage, and chemotherapy or TACE (transcatheter hepatic arterial chemoembolization) status. The stepwise Cox regression model was also conducted to identify predictive factors of HCC prognosis, with a significance level set at P < 0.050 for entering and P ≥ 0.050 for removing the respective explanatory variables. The heterogeneity between subgroups was evaluated using the chi-square-based Q-test. All the statistical analyses were carried out by the R software (Version 3.4.2, 2017-09-28; R Foundation for Statistical Computing, http://www.cran.r-project.org/).
Results
The demographic characteristics and clinical features of the 331 HCC patients were summarized previously[11,12]. Briefly, 258 of 331 HCC patients were deaths at the last time of follow-up. By univariate analysis, drinking status and chemotherapy or TACE status were significantly associated with the survival time (log-rank P = 0.006 and P ≤ 0.001 respectively). Obviously, alcohol-drinking was a risk factor of death (HR = 1.43, 95%CI = 1.11-1.84), yet Chemotherapy or TACE was a protective factor (HR = 0.39, 95%CI = 0.29-0.51).
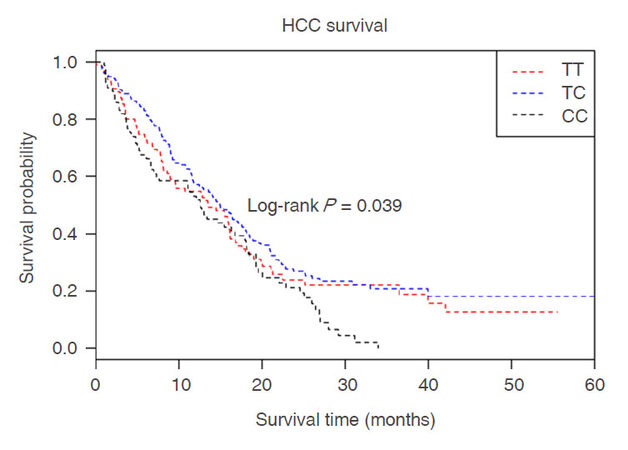
The polymorphisms of PVT1 rs4733586 and it’s association with HCC survival in different genetic models (additive models, dominant model and recessive model) were examined by log-rank test and Cox regression analyses. As shown in Table 2, patients with variant genotype CC had a higher risk of death than those with homozygous wild-type TT (adjusted HR = 1.59, 95%CI = 1.13-2.26, P = 0.008) after adjusting for age, gender, smoking status, drinking status, BCLC stage, and chemotherapy or TACE status. Furthermore, the results of the additive model analysis were also significant (adjusted HR = 1.26, 95%CI = 1.05-1.51, P = 0.012). Kaplan-Meier plot of HCC-specific overall survival by rs4733586 genotypes was shown in Figure 1. The results showed that there was a statistical significance between genotype of rs4733586 and HCC survival (log-rank P = 0.039). Stepwise Cox proportional hazard analysis was then preformed to evaluate the effect of demographic characteristics, clinical features and rs4733586 on HCC survival [Table 3]. We found that four variables(age, drinking status, chemotherapy or TACE status, PVT1 eQTL) remained in the final regression model, with a significant level of 0.050 for entering (P < 0.0001 for chemotherapy or TACE status, P = 0.0012, 0.0029 and 0.0136 for drinking status, age and rs4733586, respectively). However, in the stratified analysis [Table 4], no heterogeneity was noted among different age, gender, smoking status, drinking status, BCLC stage and chemotherapy or TACE status.
Figure 1. Kaplan-Meier plots of HCC-specific overall survival by PVT1 eQTL rs4733586 genotypes, log-rank P = 0.039. X-axis: HCC patients' survival time (months); Y-axis: HCC patients' survival probability. "Red line" denotes patients carrying homozygous wild-type TT alleles; "blue line" denotes those with heterozygous TC alleles; "black line" denotes those with variant CC alleles. HCC: hepatocellular carcinoma; PVT1: plasmacytoma variant translocation 1; eQTL: expression quantitative trait loci; TT: wild-type allele; TC: heterozygous mutant allele; CC: homozygous mutant allele
The association between polymorphisms of PVT1 genes with HCC overall survival
| Genotypes | Patients | Deaths | MST (month) | Crude HR (95% CI) | Adjusted HR (95% CI)a | Pa |
|---|---|---|---|---|---|---|
| PVT1 rs4733586 | ||||||
| TT | 95 | 73 | 13.5 | 1.00 | 1.00 | |
| TC | 153 | 115 | 14.9 | 0.85 (0.63-1.14) | 1.06 (0.78-1.44) | 0.712 |
| CC | 77 | 67 | 12.6 | 1.25 (0.90-1.75) | 1.59 (1.13-2.26) | 0.008 |
| Additive model | 1.11 (0.93-1.33) | 1.26 (1.05-1.51) | 0.012 | |||
| Dominant model | ||||||
| TT | 95 | 73 | 13.5 | 1.00 | 1.00 | |
| TC/CC | 230 | 182 | 14.3 | 0.96 (0.73-1.26) | 1.21 (0.91-1.61) | 0.191 |
| Recessive model | ||||||
| TT/TC | 248 | 188 | 14.7 | 1.00 | 1.00 | |
| CC | 77 | 67 | 12.6 | 1.39 (1.05-1.84) | 1.54 (1.15-2.05) | 0.004 |
Stepwise Cox regression analysis on HCC overall survival
| Variables | α | SE | HR | 95% CI | P |
|---|---|---|---|---|---|
| Chemotherapy/TACE | -1.2246 | 0.1540 | 0.29 | 0.22-0.40 | < 0.0001 |
| Drinking (yes vs. no) | 0.4423 | 0.1369 | 1.56 | 1.19-2.04 | 0.0012 |
| Age (≤ 53 years vs. > 53 years) | -0.4010 | 0.1348 | 0.67 | 0.51-0.87 | 0.0029 |
| rs4733586 (additive model) | 0.2263 | 0.0917 | 1.25 | 1.05-1.50 | 0.0136 |
Stratification analysis of rs4733586 genotypes and HCC overall survival
| Variables | rs4733586 (patients/deaths) | Adjusted HR | P for heterogeneity | ||
|---|---|---|---|---|---|
| TT | TC | CC | (95% CI)a | ||
| Age, years | 0.146 | ||||
| ≤ 53 | 49/38 | 83/65 | 37/32 | 1.05 (0.82-1.33) | |
| > 53 | 46/35 | 70/50 | 40/35 | 1.37 (1.05-1.79) | |
| Gender | 0.485 | ||||
| Male | 77/59 | 138/103 | 65/56 | 1.18 (0.97-1.44) | |
| Female | 18/14 | 15/12 | 12/11 | 1.40 (0.91-2.17) | |
| Smoking | 0.721 | ||||
| Never | 38/31 | 57/37 | 24/22 | 1.14 (0.85-1.56) | |
| Ever | 57/42 | 96/78 | 53/45 | 1.23 (0.98-1.54) | |
| Drinking | 0.634 | ||||
| Never | 36/28 | 68/46 | 22/20 | 1.12 (0.80-1.55) | |
| Ever | 59/45 | 85/69 | 55/47 | 1.23 (0.99-1.53) | |
| BCLC stage | 0.071 | ||||
| Stage B | 89/68 | 142/106 | 69/61 | 1.29 (1.07-1.55) | |
| Stage C | 6/5 | 11/9 | 8/6 | 0.56 (0.23-1.36) | |
| Chemotherapy/TACE | 0.135 | ||||
| No | 33/26 | 32/26 | 25/25 | 1.27 (0.94-1.71) | |
| Yes | 62/47 | 121/89 | 52/42 | 0.96 (0.77-1.19) | |
Discussion
In this present case cohort study, we genotyped the PVT1 eQTL (rs4733586) among 331 HCC patients and shed light on that the variants of SNP were significantly associated with poor prognosis in HCC.
Several studies have shown that some locus located in PVT1 had potential risks to cancer.For example,one genome-wide association study identified a locus (rs1561927) at 8q24.21 that located 455 Kb telomeric of PVT1 associated with pancreatic cancer risk[15]. In a comprehensive genome-wide analysis, the authors identified lncRNA PVT1 that may be involved in HCC cells metastasis by comparing lncRNAs expression profiles[16]. Therefore, it is reasonable to believe that the key locus on the lncRNA PVT1 may be associated with the progress of HCC.
Since thousands of new lncRNAs have been explored in the ENCODE project and RNA-seq analysis, the genetic variation and biological function of lncRNAs are becoming hot topics in cancer[12]. SNP rs4733586 was identified as an eQTL for PVT1 using bioinformatics analysis. PVT1 oncogene encodes a long noncoding RNA and maps to chromosome 8q24.21[17]. The well-characterized myelocytomatosis (MYC) oncogene also resides in the 8q24.21 region[18], and PVT1 is located downstream of MYC in this chromosomal region[9]. Moreover, PVT1 has been shown to be important for expression of MYC in tumors[19]. MYC activation may influence cancer immunoediting through the suppression of immune surveillance against tumor cells. During tumor progression, high MYC expression results in increased expression of CD47 and PD-L1, suppressing both the innate and the adaptive immune response and favoring tumor growth[20]. Previous studies had shown that there was a significant relationship between PVT1 overexpression and poor overall survival of patients with gastric cancer, gynecology cancer and lung cancer[7]. Ding et al.[21] found that the relative expression levels of PVT1 were significantly higher in cancerous tissues compared with the corresponding non-cancerous tissues. Other research group demonstrated that PVT1 promotes cell proliferation, cell cycling, and the acquisition of stem cell-like properties in HCC cells by stabilizing NOP2 protein, and HCC patients with high PVT1 expression had a poor prognosis[22]. All these conclusions can be consistent with the results of this study.
However, there are several limitations of the study that need to be addressed in further studies. Firstly, the further verification needs to be conducted. A series of large-scale studies are needed to verify the associations between the eQTL in PVT1 and the HCC prognosis. Secondly, there was few biological functional experiments conducted to provide additional evidence.
In conclusion, it was the first study to examine the association of PVT1 eQTL with HCC prognosis. We found that rs4733586 might be served as a susceptibility marker for HCC survival.
Declarations
Authors’ contributionsSNP selection and genotype: Ge ZJ , Yu CX
Data acquisition: Song C, Pu ZN
Statistical analysis: Tian T, Song C
Manuscript preparation: all authors
Critical revision and finalizing of the manuscript: Song C, Hu ZB
Availability of data and materialsThe datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Financial support and sponsorshipThis work was supported by grants from the National Key Basic Research Program Grant (2013CB911400); Cheung Kong Scholars Programme of China; Jiangsu Specially-Appointed Professor project; the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine) and Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067). The funders had no role in the study design, data collection, data analysis and interpretation, writing of the report, or the decision to submit the article for publication.
Conflicts of interestAll authors declare that there are no conflicts of interest.
Ethical approval and consent to participateThis study was authorized by the local institutional review board at Nanjing Medical University. All the participants gave written informed consent.
Consent for publicationNot applicable.
Copyright© The Author(s) 2018.
REFERENCES
1. Wheeler DA. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327-41.e23.
4. Waghray A, Murali AR, Menon KN. Hepatocellular carcinoma: from diagnosis to treatment. World J Hepatol 2015;7:1020-9.
5. Chen G, Li X, He G, Yu Z, Luo J, He J, Huang Z. Low expression of GNAI3 predicts poor prognosis in patients with HCC. Int J Clin Exp Med 2015;8:21482-6.
6. Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int 2013;2013:251098.
7. Zhu S, Shuai P, Yang C, Zhang Y, Zhong S, Liu X, Chen K, Ran Q, Yang H, Zhou Y. Prognostic value of long non-coding RNA PVT1 as a novel biomarker in various cancers: a meta-analysis. Oncotarget 2017;8:113174-84.
8. Du M, Yuan T, Schilter KF, Dittmar RL, Mackinnon A, Huang X, Tschannen M, Worthey E, Jacob H, Xia S, Gao J, Tillmans L, Lu Y, Liu P, Thibodeau SN, Wang L. Prostate cancer risk locus at 8q24 as a regulatory hub by physical interactions with multiple genomic loci across the genome. Hum Mol Genet 2015;24:154-66.
9. Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in cancer: a review and meta-analysis. Clin Chim Acta 2017;474:1-7.
10. Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H, Hu Z. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 2012;55:1426-31.
11. Xie K, Liu J, Zhu L, Liu Y, Pan Y, Wen J, Ma H, Zhai X, Hu Z. A potentially functional polymorphism in the promoter region of let-7 family is associated with survival of hepatocellular carcinoma. Cancer Epidemiol 2013;37:998-1002.
12. Ma S, Yang J, Song C, Ge Z, Zhou J, Zhang G, Hu Z. Expression quantitative trait loci for PAX8 contributes to the prognosis of hepatocellular carcinoma. PLoS One 2017;12:e0173700.
14. Yin J, Wen J, Hang D, Han J, Jiang J, Song C, Liu Y, Liu J, Liu L, Zhu L, Chen J, Zhai X, Xie S, Hu Z, Shen H, Dai M, Li N. Expression quantitative trait loci for CARD8 contributes to risk of two infection-related cancers--hepatocellular carcinoma and cervical cancer. PLoS One 2015;10:e0132352.
15. Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, Beane-Freeman L, Bracci PM, Buring J, Canzian F, Duell EJ, Gallinger S, Giles GG, Goodman GE, Goodman PJ, Jacobs EJ, Kamineni A, Klein AP, Kolonel LN, Kulke MH, Li D, Malats N, Olson SH, Risch HA, Sesso HD, Visvanathan K, White E, Zheng W, Abnet CC, Albanes D, Andreotti G, Austin MA, Barfield R, Basso D, Berndt SI, Boutron-Ruault MC, Brotzman M, Buchler MW, Bueno-de-Mesquita HB, Bugert P, Burdette L, Campa D, Caporaso NE, Capurso G, Chung C, Cotterchio M, Costello E, Elena J, Funel N, Gaziano JM, Giese NA, Giovannucci EL, Goggins M, Gorman MJ, Gross M, Haiman CA, Hassan M, Helzlsouer KJ, Henderson BE, Holly EA, Hu N, Hunter DJ, Innocenti F, Jenab M, Kaaks R, Key TJ, Khaw KT, Klein EA, Kogevinas M, Krogh V, Kupcinskas J. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 2014;46:994-1000.
16. Cui H, Zhang Y, Zhang Q, Chen W, Zhao H, Liang J. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer Med 2017;6:2932-41.
17. Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, Xia R, Wan L, Sun M, Wang ZX, De W, Zhang ZH. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer 2015;14:82.
18. Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, Essig J, Otto GM, O'Sullivan MG, Largaespada DA, Schwertfeger KL, Marahrens Y, Kawakami Y, Bagchi A. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014;512:82-6.
19. Bronson PG, Chang D, Bhangale T, Seldin MF, Ortmann W, Ferreira RC, Urcelay E, Pereira LF, Martin J, Plebani A, Lougaris V, Friman V, Freiberger T, Litzman J, Thon V, Pan-Hammarstrom Q. Common variants at PVT1, ATG13-AMBRA1, AHI1 and CLEC16A are associated with selective IgA deficiency. Nat Genet 2016;48:1425-9.
20. Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227-31.
21. Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng C, Cheng S, Xie H, Zhou L, Wu J, Zheng S. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett 2015;9:955-63.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Tian T, Song C, Pu ZN, Ge ZJ, Yu CX, Liu JB, Hu ZB. Expression quantitative trait loci for PVT1 contributes to the prognosis of hepatocellular carcinoma. Hepatoma Res 2018;4:27. http://dx.doi.org/10.20517/2394-5079.2018.24
AMA Style
Tian T, Song C, Pu ZN, Ge ZJ, Yu CX, Liu JB, Hu ZB. Expression quantitative trait loci for PVT1 contributes to the prognosis of hepatocellular carcinoma. Hepatoma Research. 2018; 4: 27. http://dx.doi.org/10.20517/2394-5079.2018.24
Chicago/Turabian Style
Tian, Ting, Ci Song, Zhe-Ning Pu, Zi-Jun Ge, Cheng-Xiao Yu, Ji-Bin Liu, Zhi-Bin Hu. 2018. "Expression quantitative trait loci for PVT1 contributes to the prognosis of hepatocellular carcinoma" Hepatoma Research. 4: 27. http://dx.doi.org/10.20517/2394-5079.2018.24
ACS Style
Tian, T.; Song C.; Pu Z.N.; Ge Z.J.; Yu C.X.; Liu J.B.; Hu Z.B. Expression quantitative trait loci for PVT1 contributes to the prognosis of hepatocellular carcinoma. Hepatoma. Res. 2018, 4, 27. http://dx.doi.org/10.20517/2394-5079.2018.24
About This Article
Copyright
Data & Comments
Data

 Cite This Article 1 clicks
Cite This Article 1 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.